2026
146. Sang, M., Yang, Q., Guo, J., Feng, P., Gao, Y., Ma, W., Li, S., Metsä-Ketelä, M., Zhang, W.* Functional Plasticity of Methyltransferases in Anthracycline Biosynthesis: A Single Residue Reversal between Decarboxylation and Hydroxylation. ACS Catal. 2026, doi.org/10.1021/acscatal.5c07819. | PDF
2025
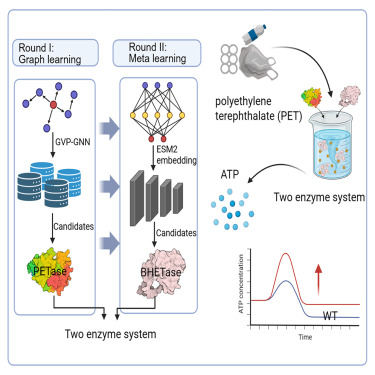
145. Li, A. #, Song, Y. #, Hu, J. #, Zhang, H., Li, Z., Tang, J., Huang, H., Li, S. *, Li, X. * Integrating graph learning and meta-learning to enhance PET hydrolase activity at elevated temperatures. Cell Rep. Phys. Sci. 2025, 103037. | PDF
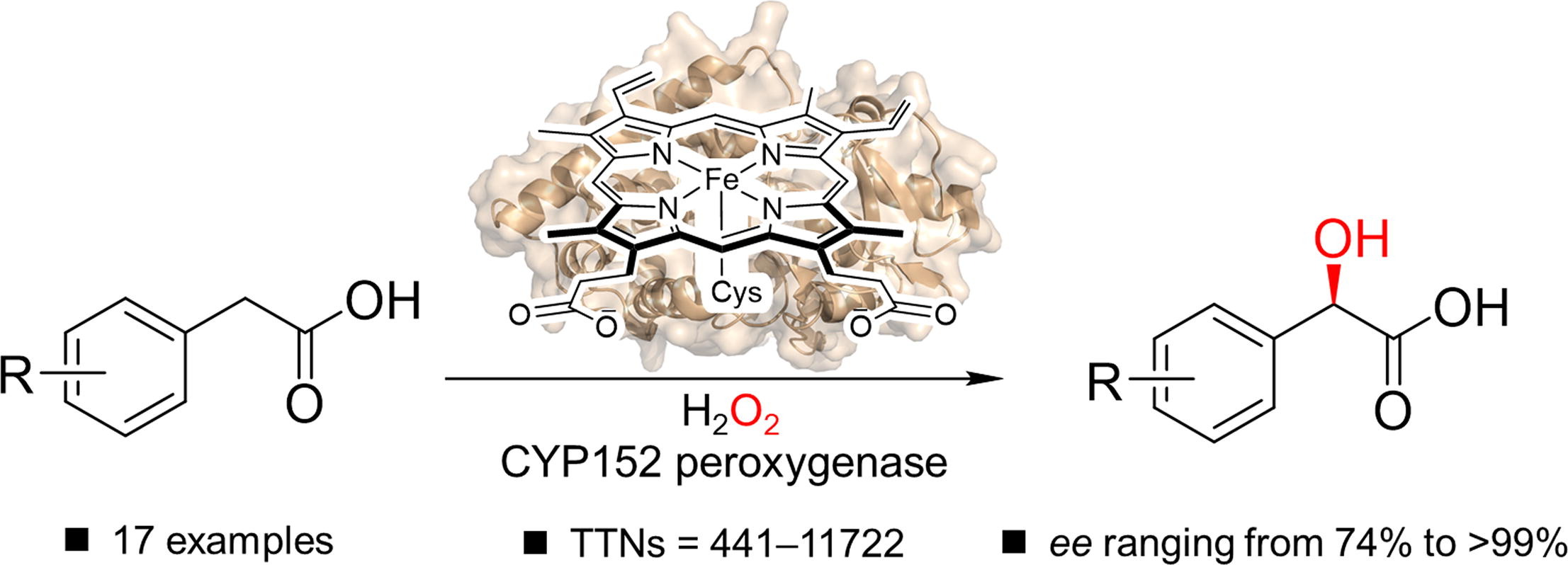
144. Li, Z. #, Jiang, Y. #, Li, N., Li, S.* Efficient and enantioselective biosynthesis of (R )-mandelic acid derivatives using CYP152 peroxygenases. Sci. Bull. 2025, doi.org/10.1016/j.scib.2025.10.031. | PDF
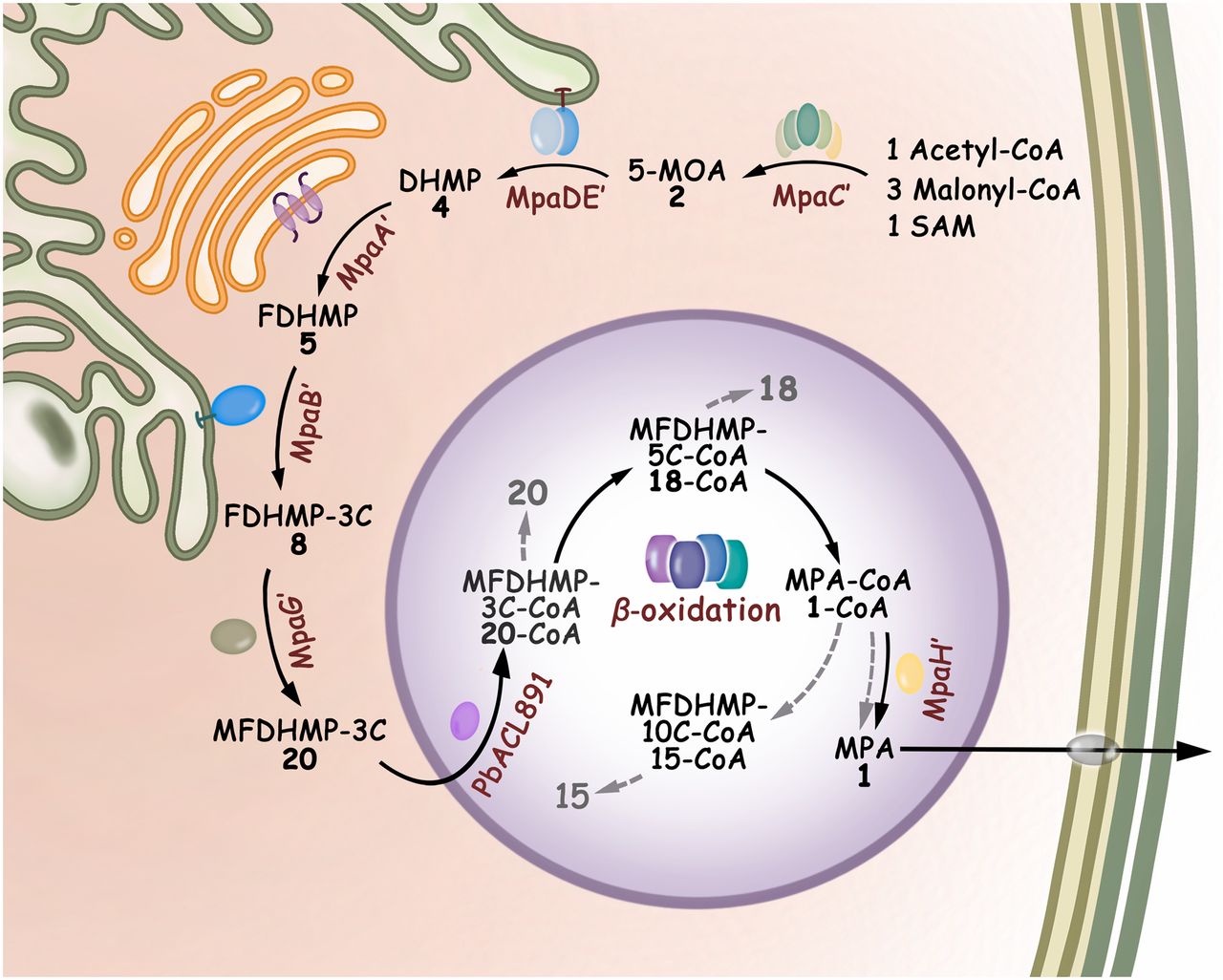
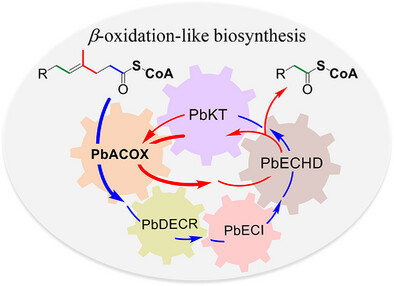
143. Fan, B., Liu, Y., Chi, L., Yang, C., Li, S. *, Zhang, W. * Enhancing the 𝜷-Oxidation-Like Pathway for the OptimalProduction of the Immunosuppressant Mycophenolic Acid. Adv. Sci. 2025, e08826. | PDF
142. Guo, J. #, Zhang, Q. #, Shen, Y. #, Cheng, F., Sang, M., Wang, X., Pan, Y., Liu, M., Yu, H., Hu, B., Wang, S., Zheng, L., Geng, C., Yang, C., Luo, L., Zhang, G., Du, L., Li, Y., Zhang, W., Zhang, Y.*, Wang, B.*, Li, S.*, Zhang, X.* Deciphering the Thiolactonization Mechanism in Thiolactomycin Biosynthesis. J. Am. Chem. Soc. 2025, doi.org/10.1021/jacs.4c14296.| PDF
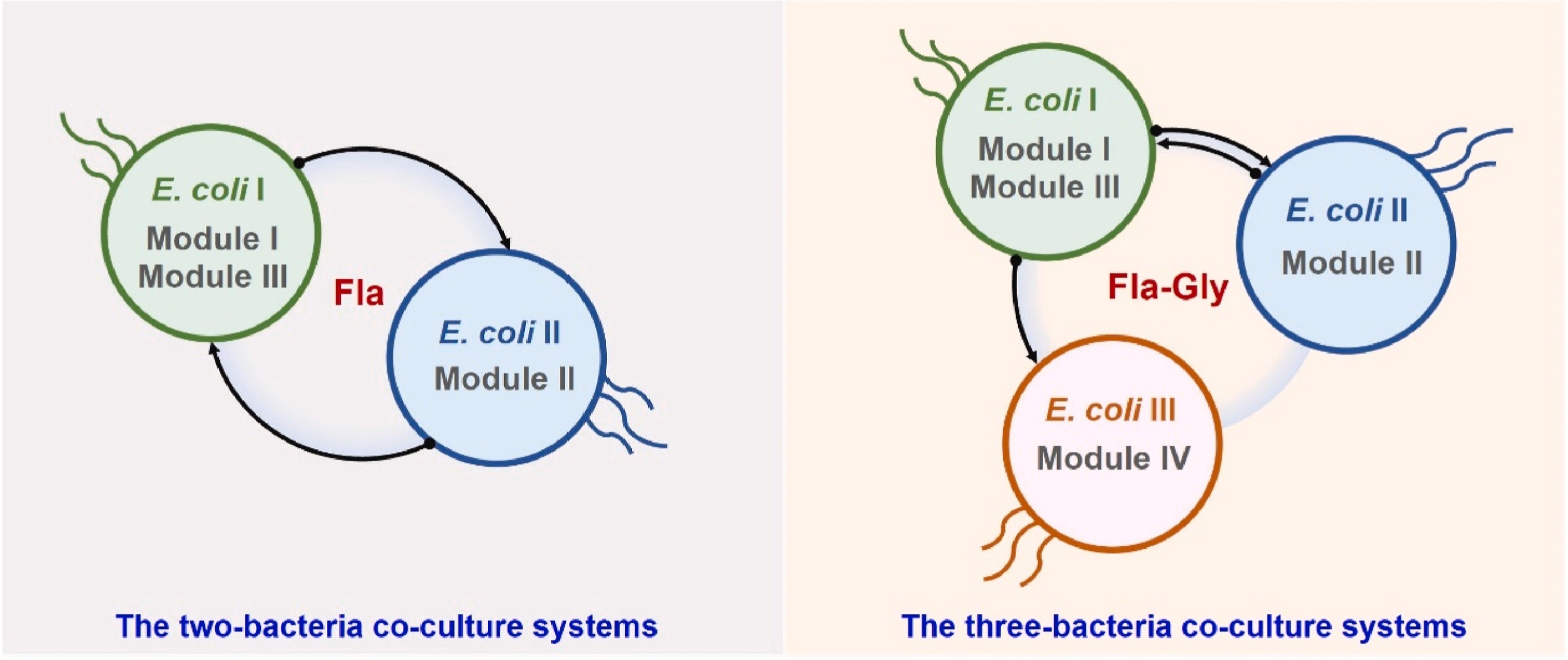
141. Qiu, Z., Han, Y., Li, J., Ren, Y., Liu, X., Li, S. *, Zhao, G. *, Du, L. * Metabolic division engineering of Escherichia coli consortia for de novo biosynthesis of flavonoids and flavonoid glycosides. Metab. Eng. 2025, 89, 60-75. | PDF
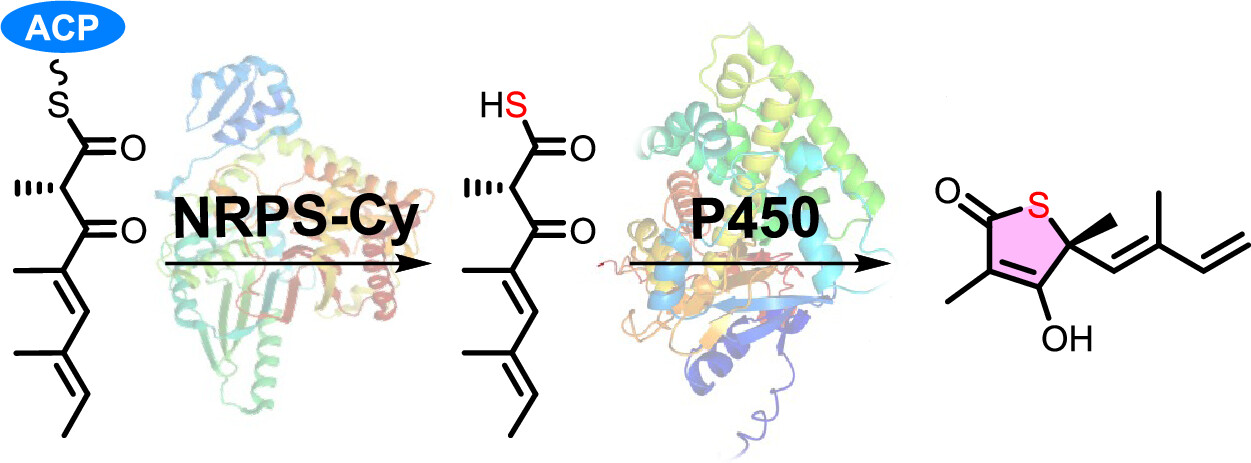
140. Wang, X., Tian, X., Guo, J., Cheng, F., Liu, M., Zheng, S., Feng, Y., Lv, Y., Li, Y., Li, S., Zhang, X.* Carboxylic Acid Tailoring in Thioquinolobactin Biosynthesis. J. Nat. Prod. 2025, doi.org/10.1021/acs.jnatprod.5c00331.| PDF
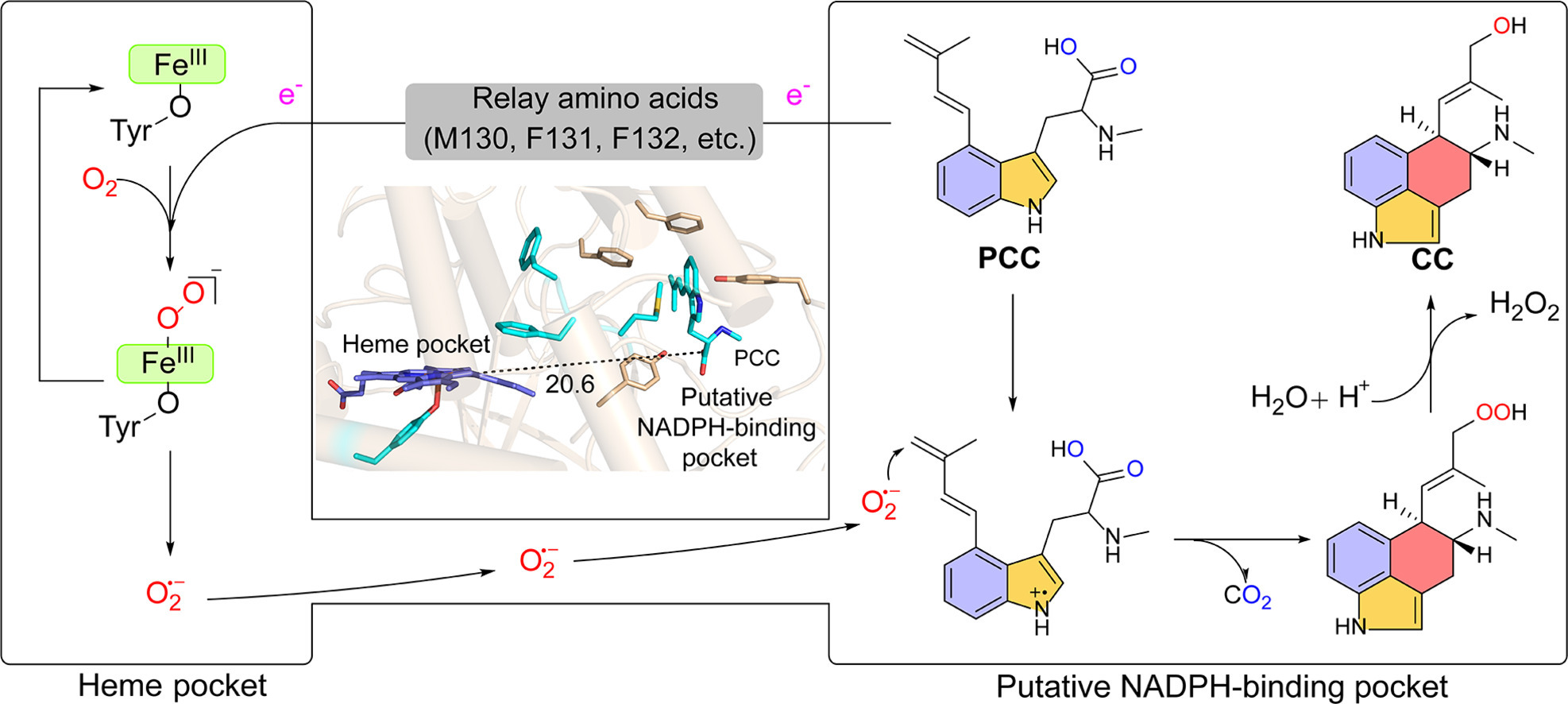
139. Jiang, Y. #, Li, Z. #, Li, S. * Superoxide-mediated O2 activation drives radical cyclization in ergot alkaloid biosynthesis. Eng. Microbiol. 2025, 5, 100207.| PDF
138. Zhang, C.#, Zhou, J.#, Wang, W.#, Liu, M.#, Li, Y., Sheng, X.*, Du, L.*, Li, S.* Oxidative Electrocyclization Assembles 2,2-Dimethyl-2H -pyran in Fungal Indole Alkaloids. Angew. Chem. Intl. Ed. 2025, e202506276. | PDF
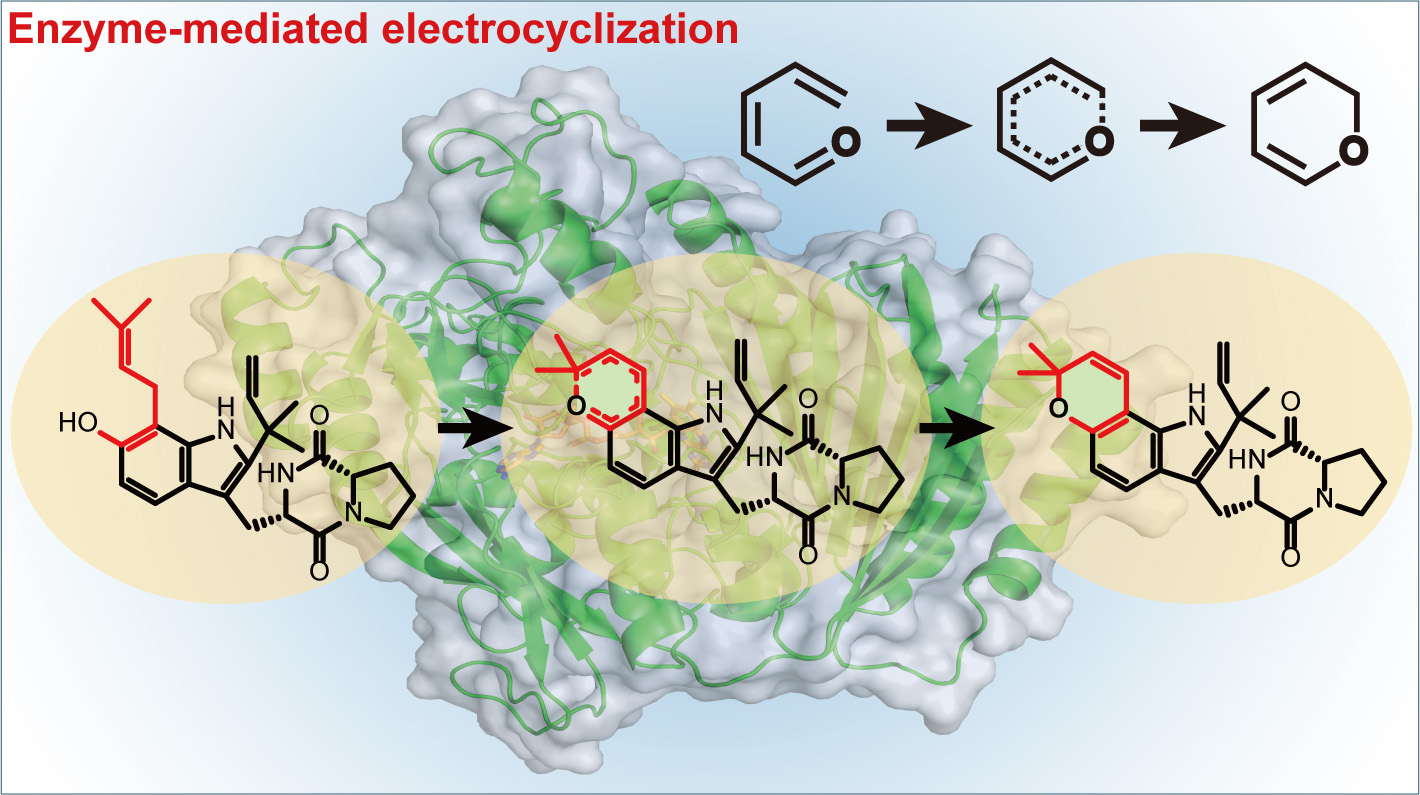
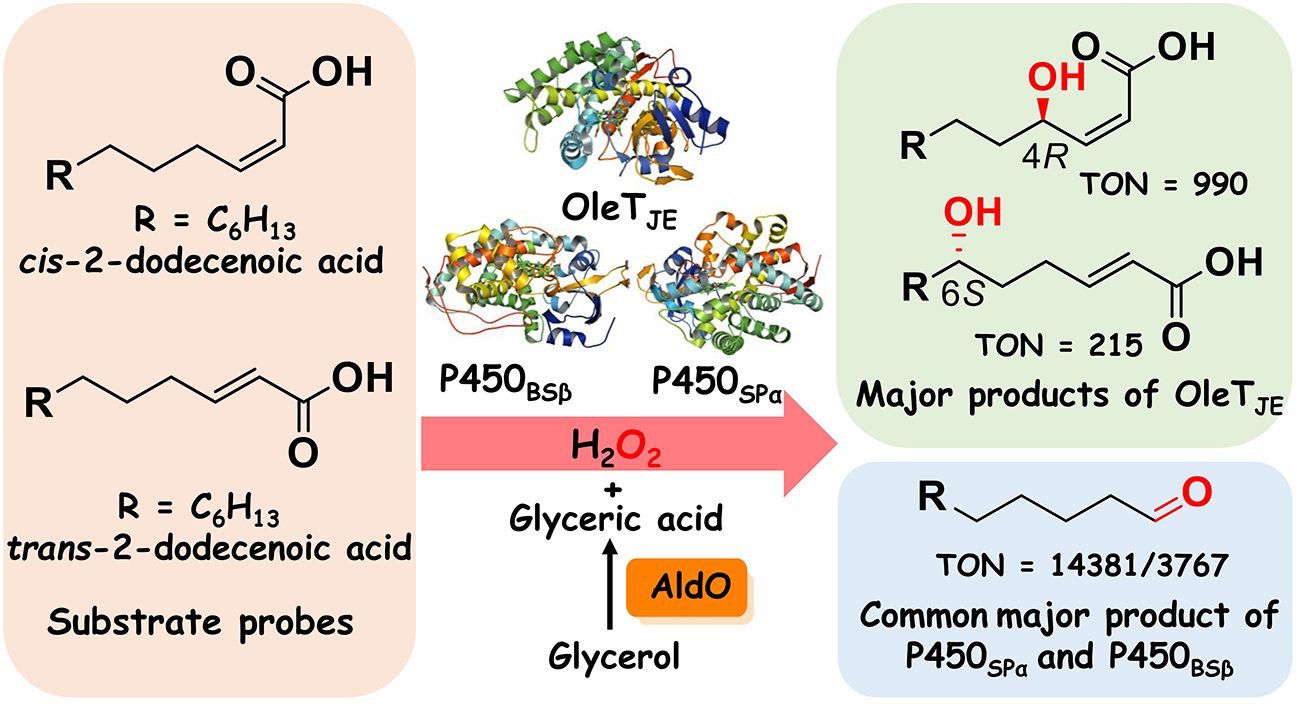
137. Jiang, Y. #, Gong, P. #, Li, Z. #, Li, Z., Li, Y., Wang, B., Huang, H.*, Peng, W.*, Gao, X.*, Li, S.* Unexpected Activities of CYP152 Peroxygenases towards Non-carboxylic Substrates Reveal Novel Substrate Recognition Mechanism and Catalytic Versatility. Angew. Chem. Intl. Ed. 2025, e202506614. | PDF
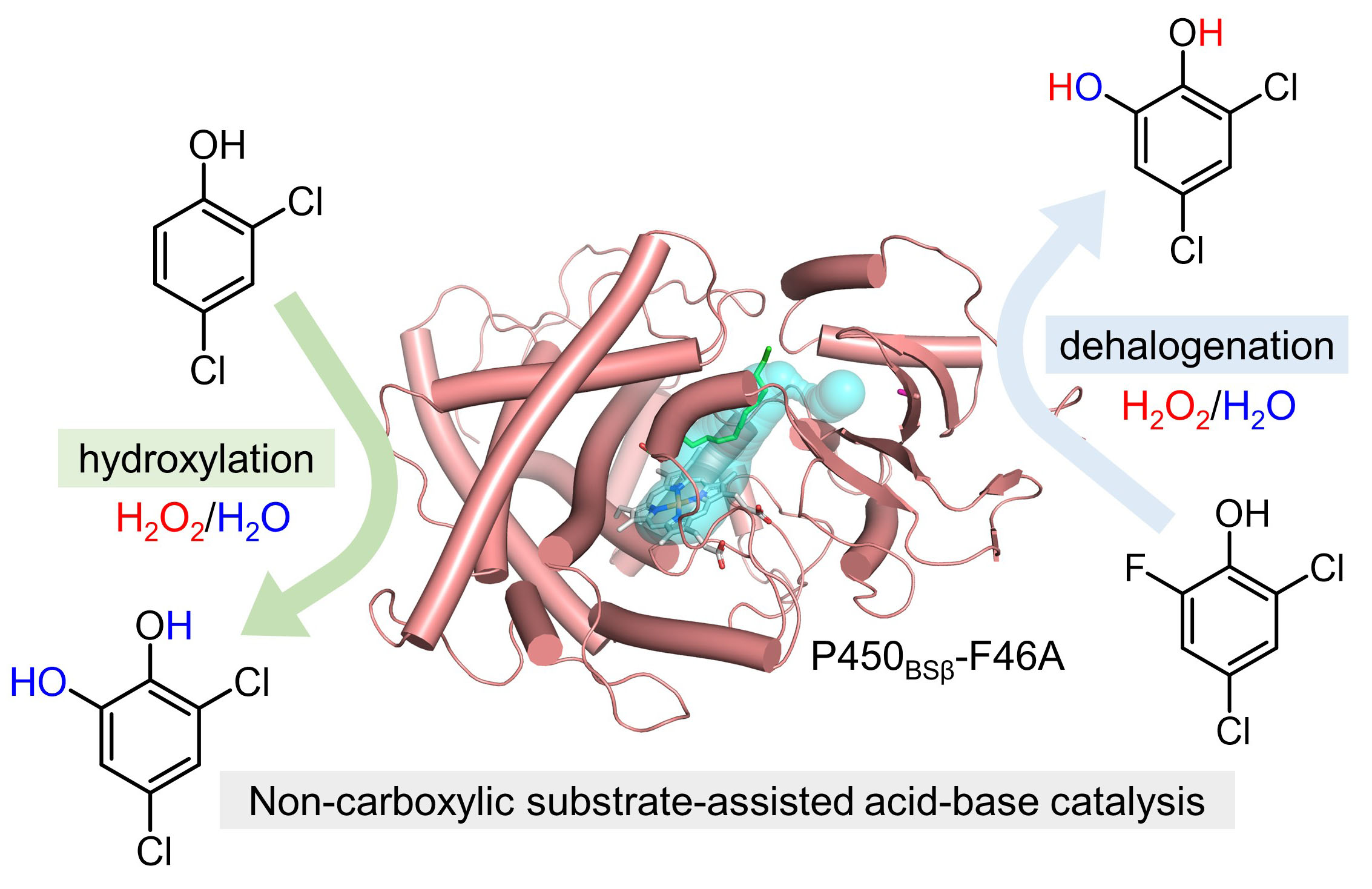
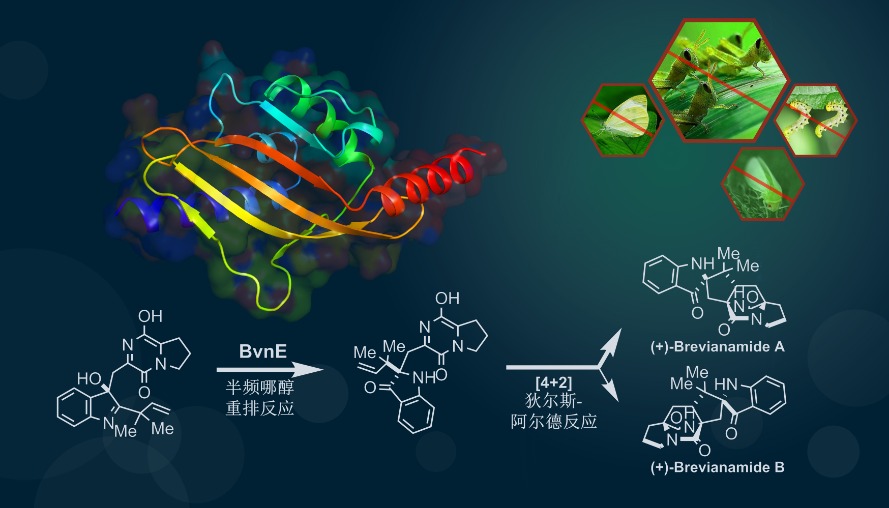
136. Du, L.#, Dian, L.#, Newmister, Sean A., Xia, Y., Luo, G., Sherman, David H.*, Li S.* The Mutually Inspiring Biological and Chemical Synthesis of Fungal Bicyclo[2.2.2]diazaoctane Indole Alkaloids. Chem. Rev. 2025, doi.org/10.1021/acs.chemrev.4c00250.| PDF
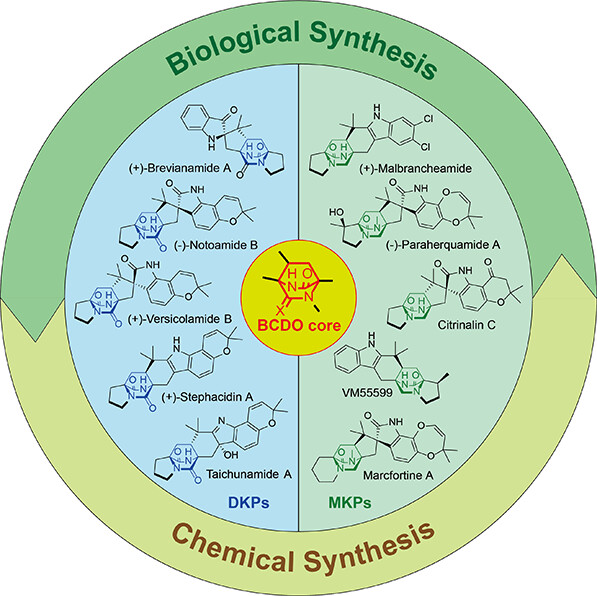
135. Sun, T. #, Liu, M. #, Li, S., Guo, J., Liu, L., Zhang, L., Ma L.*, Li, S.* Cytochrome P450-Catalyzed Tetrahydrofuran Formation via Dual Pathways in Avermectin Biosynthesis. ACS Catal. 2025, 15, 3295-3305.| PDF
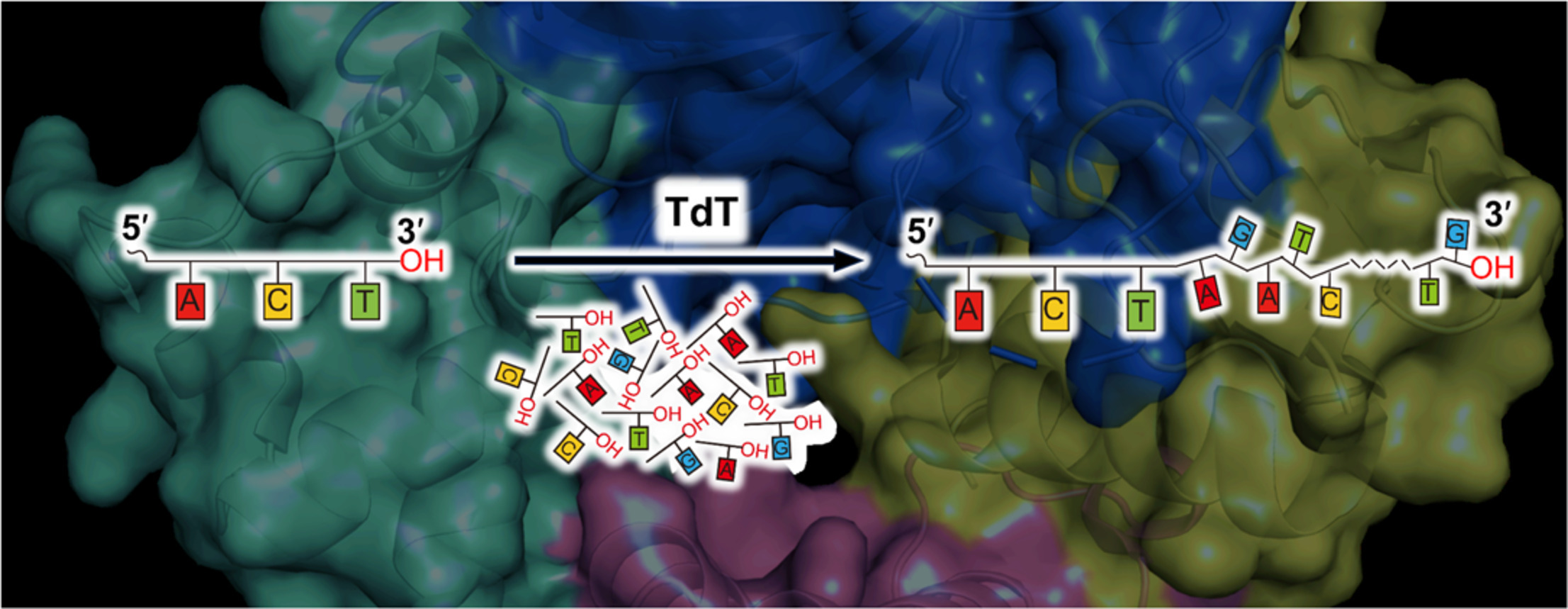
134. Zhang, C., Subthain, H., Guo, F., Fang, P., Zheng, S., Shen, M., Jiang, X., Gao, Z., Meng, C. *, Li, S.*, Du, L. * Terminal deoxynucleotidyl transferase: Properties and applications. Eng. Microbiol. 2025, 5, 100179. | PDF
133. Li, W. #, Zheng, S. #, Xu, X., He, J., Jiao, X., Wang, M., Hu, W., Li, S., Jiang, X., Lim, B., Shao, F., Gao, X. * A conserved adaptor orchestrates co-secretion of synergistic type VI effectors in gut Bacteroidota. Cell Host Microbe 2025, 33, 1901-1915. | PDF
132. Liu, K., Ma, Z., Deng, M., Ma, T., Li, S., Qi, Q., Dian, L.* Photo-induced solvent-enabled catalyst-/additive free selective C(α)–C(β) bond cleavage of β-O-4 ketone lignin model compounds at room temperature. Green Chem. 2025, 27, 2689-2695.| PDF
131. Zhang, G., Xu, Z., Han, B., Ji, Y., Li, S., Zhou, M. *, Cao, M. *, Yu, X. *, Liu, L. * Iron-Catalyzed Site-Selective Bromination of Benzylic C(sp3)−HBonds. Org. Lett. 2025, 27, 3720-3724.| PDF
130. Feng, B. #, Li, Y. #, Xu, B. #, Liu, H., Steenwyk, L. J., David, T. K., Tian, X., Gonçalves, C., Opulente, A. D., LaBella, L. A., Harrison, C. M., Wolters, F. J., Shao, S., Chen, Z., Fisher, J. K., Groenewald, M., Hittinger, T. C., Shen, X., Li, S., Rokas, A*., Zhou, X. *, Li, Y. * Unique trajectory of gene family evolution fromgenomic analysis of nearly all known species in anancient yeast lineage. Mol. Syst. Biol. 2025, doi. org/10.1038/s44320-025-00118-0. | PDF
2024
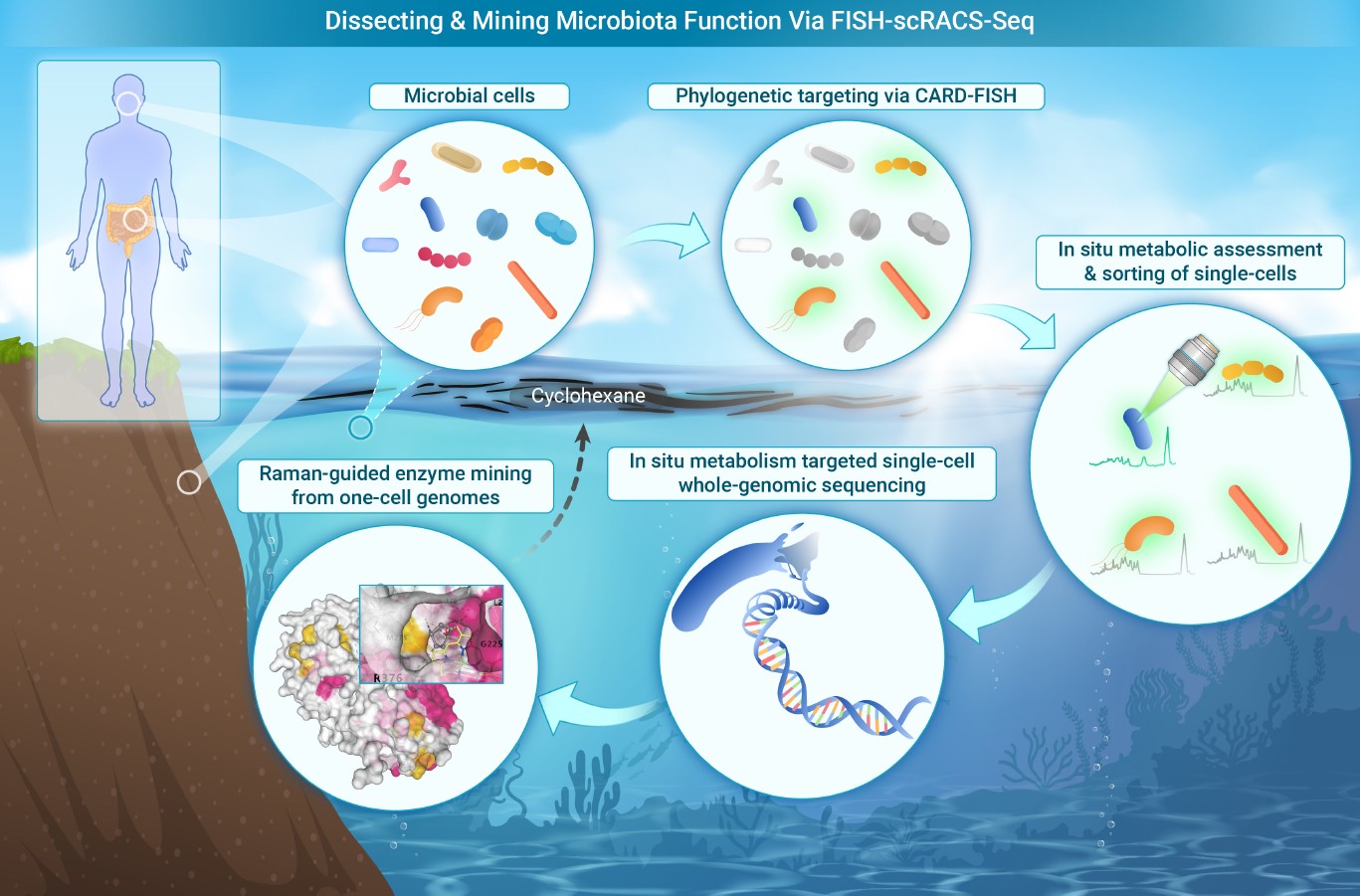
129. Jing, X.#, Gong, Y.#, Diao, Z.#, Ma, Y.#, Meng, Y., Chen, J., Ren, Y., Liang, Y., Li, Y., Sun, W., Zhang, J., Ji, Y., Li, S., Ma, B., Cui, Z.*, Ma, L.*, Xu, J.* Phylogeny-metabolism dual-directed single-cell g enomics for issecting and mining ecosystem function by FISH-scRACS-seq. The Innovation 2024, doi.org/10.1016/j.xinn.2024.100759. | PDF
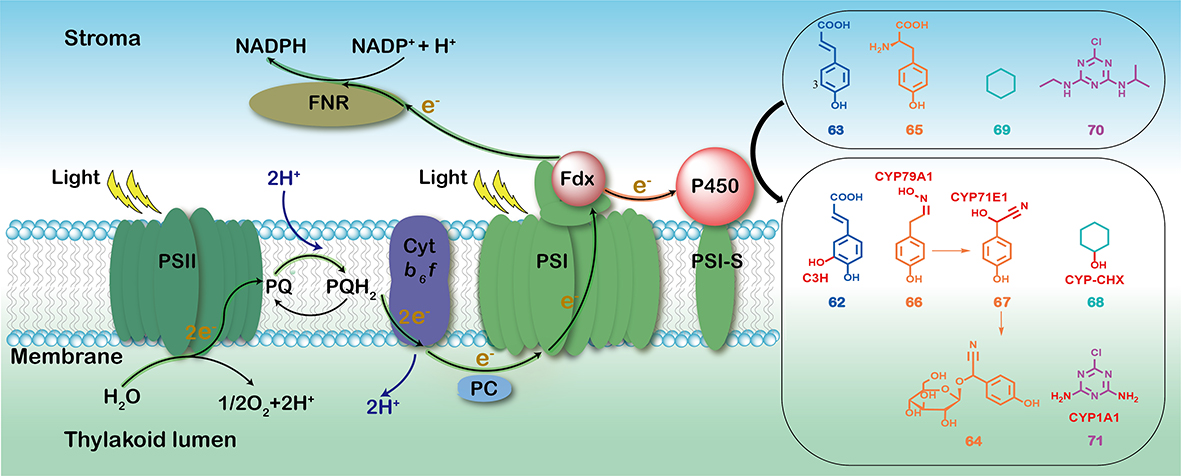
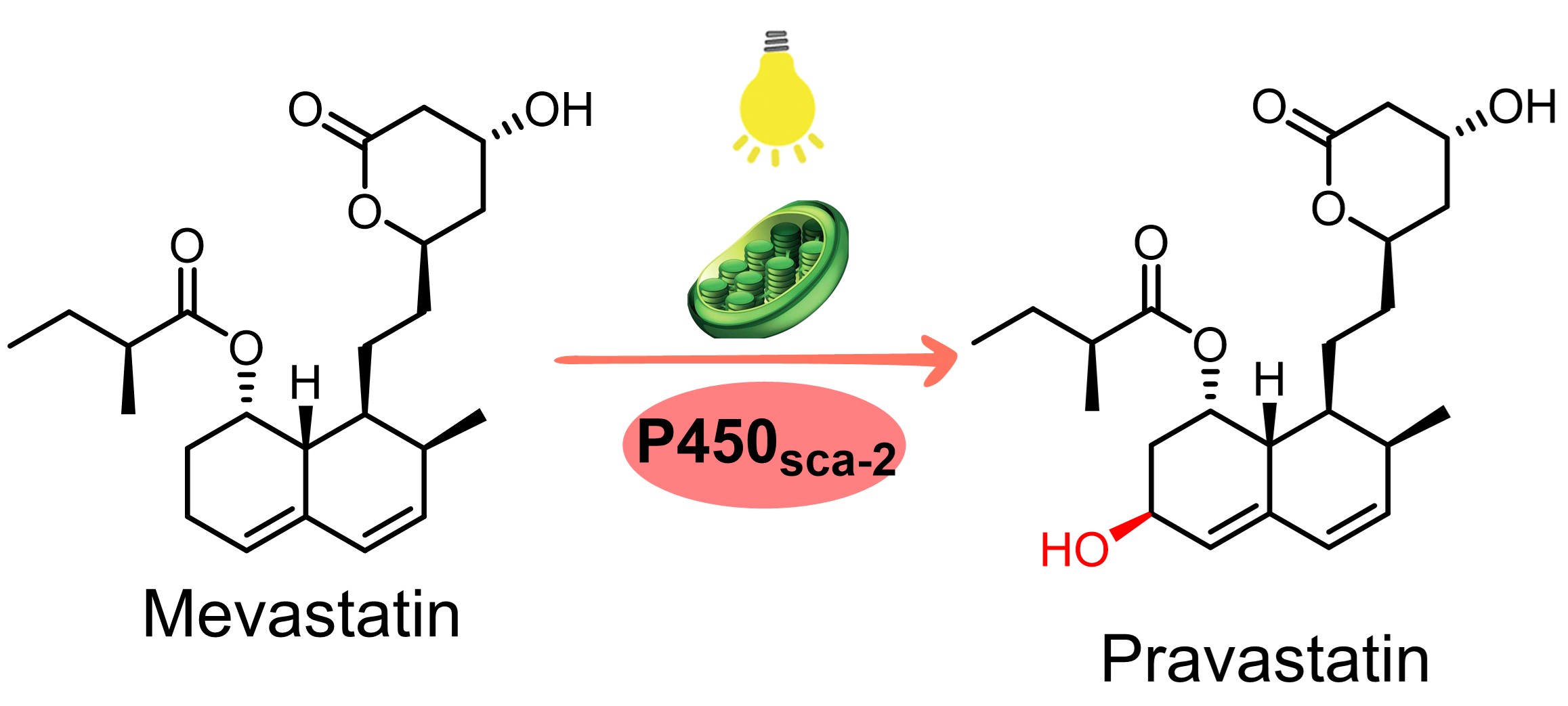
128. Zheng, S. #, Gao, Z. #, Jiang, Y., Guo, J., Cheng, F., Wang, X., Yu, H., Hu, B., Meng, C.*, Li, S.*, Zhang, X.* Light-driven enzymatic conversion of mevastatin into pravastatin by coupling P450sca-2 with cyanobacterial photosynthetic system. Green Synth. Catal. 2024, doi.org/10.1016/j.gresc.2024.07 .001. | PDF
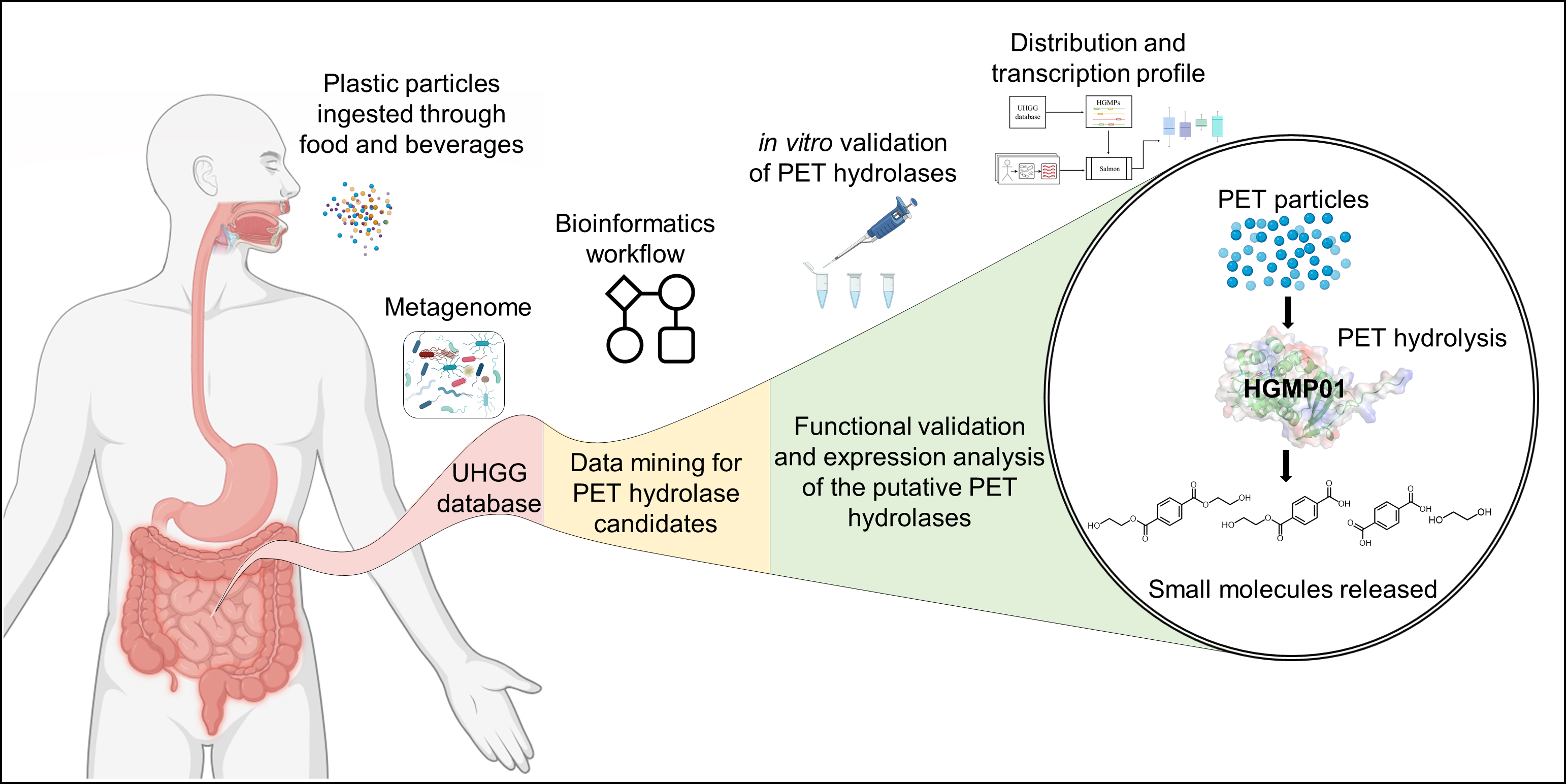
127. Zhang, G., Du, J., Zhang, C., Zhao, Z., Chen, Y., Liu, M., Chen, J., Fan, G., Ma, L.*, Li, S.*, Liu, K.* Identification of a PET hydrolytic enzyme from the human gut microbiome unveils potential plastic biodegradation in human digestive tract. Int. J. Biol. Macromol. 2024, 283, 137732.| PDF
126. Wang, A., Wang, Y., You, Y., Huang, Z., Zhang, X., Li, S. *, Chen, H. * One-pot Biocatalytic Conversion of Chemically Inert Hydrocarbons into Chiral Amino Acids through Internal Cofactor and H2O2 Recycling. Angew. Chem. Intl. Ed. 2024, 63, 48, e202410260. | PDF

125. Chen, J. #, Jia, Y. #, Sun, Y. #*, Liu, K. #, Zhou, C., Liu, C., Li, D., Liu, G., Zhang, C., Yang, T., Huang, L., Zhuang, Y., Wang, D., Xu, D., Zhong, Q., Guo, Y., Li, A., Seim, I., Jiang, L., Wang, L., Lee, M. Y. S., Liu, Y., Wang, D., Zhang, G., Liu, S., Wei, X., Yue, Z., Zheng, S., Shen, X., Wang, S., Qi, C., Chen, J., Ye, C., Zhao, F., Wang, J., Fan, J., Li, B., Sun, J., Jia, X., Xia, Z., Zhang, H., Liu, J., Zheng, Y., Liu, X., Wang, J., Yang, H., Kristiansen, K., Xu, X., Mock, T. *, Li, S. *, Zhang, W. *, Fan, G. * Global marine microbial diversity and its potential in bioprospecting. Nature 2024, 633, 371-379. | PDF

124. Tang, D., Zheng, X., Zhao, Y., Zhang, C., Chen, C., Chen, Y., Du, L. *, Liu, K. *, Li, S. * Engineered Microbial Consortium for De Novo Production of Sclareolide. J. Agric. Food Chem. 2024, 72, 36, 19977-19984. | PDF

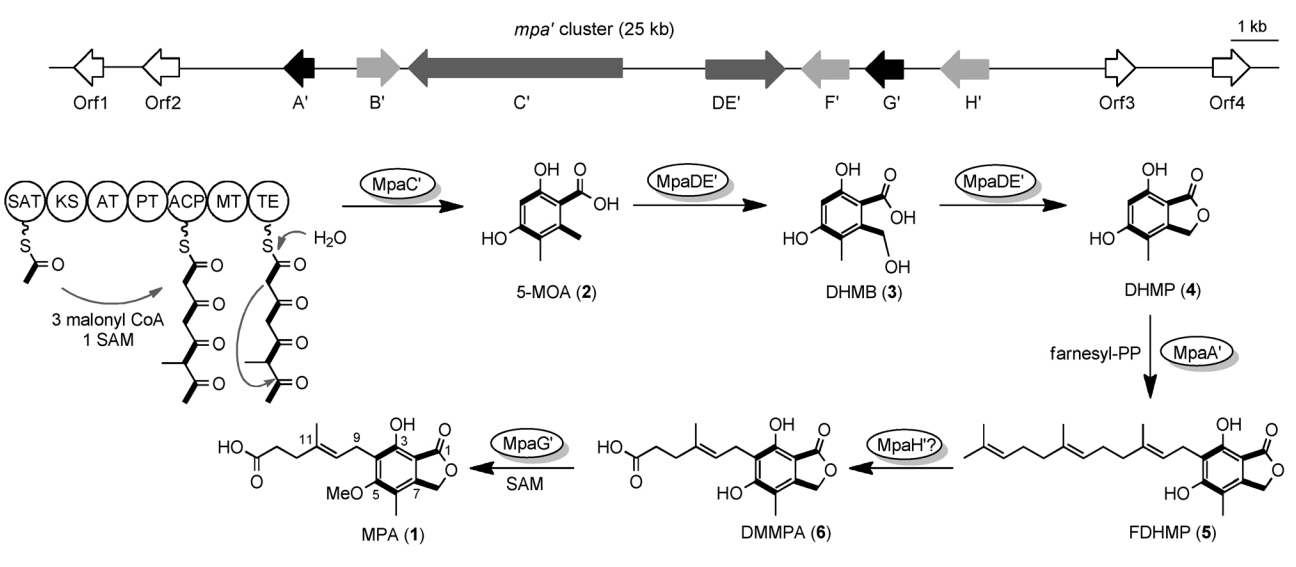
123. You, C. #, Pan, Y. #, Liu, R., Li, S. *, Feng, Y. * Structural basis for substrate flexibility of the O-methyl transferase MpaG' involved in mycophenolic acid biosynthesis. Protein Sci. 2024, 33(9), e5144. | PDF

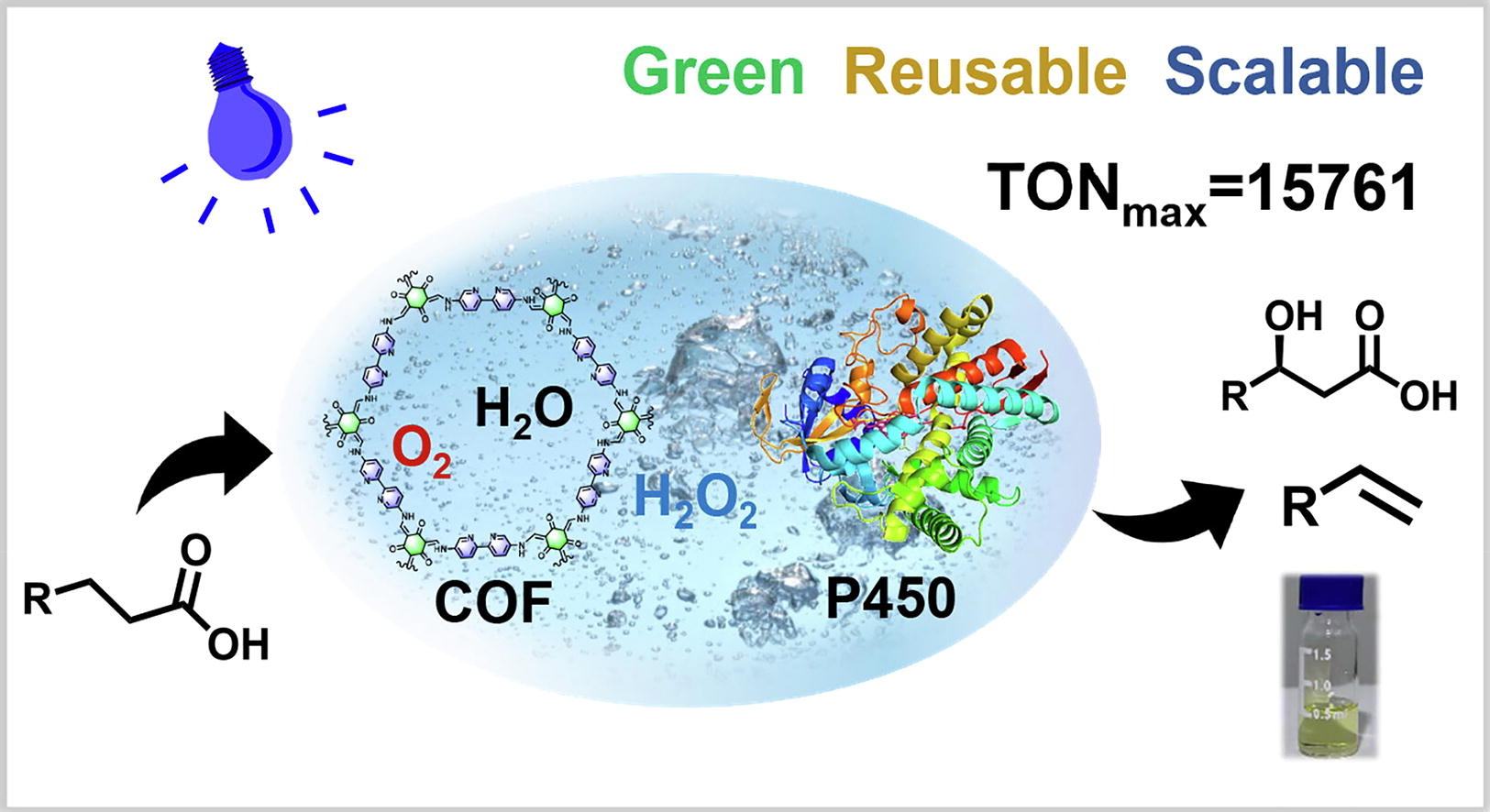
122. Jiang, Y. #, Li, P. #, Li, Z., Zhang, B. *, Li, S. * Photoenzymatic synthesis of 1-alkenes and hydroxyl fatty acids by cascading a COF photocatalyst and P450 peroxygenases. Sci. Bull. 2024,69(21), 3350-3354.| PDF
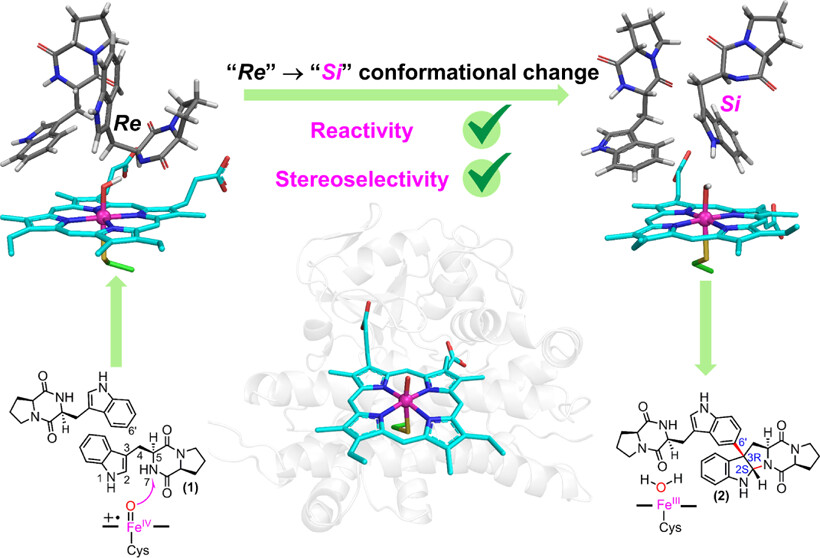
121. Zhou, T., Feng, J., Wang, Y., Li, S.*, Wang, B.* Substrate Conformational Switch Enables the Stereoselective Dimerization in P450 NascB: Insights from Molecular Dynamics Simulations and Quantum Mechanical/Molecular Mechanical Calculations. J. Am. Chem. Soc. Au 2024, 4, 1591-1604.| PDF
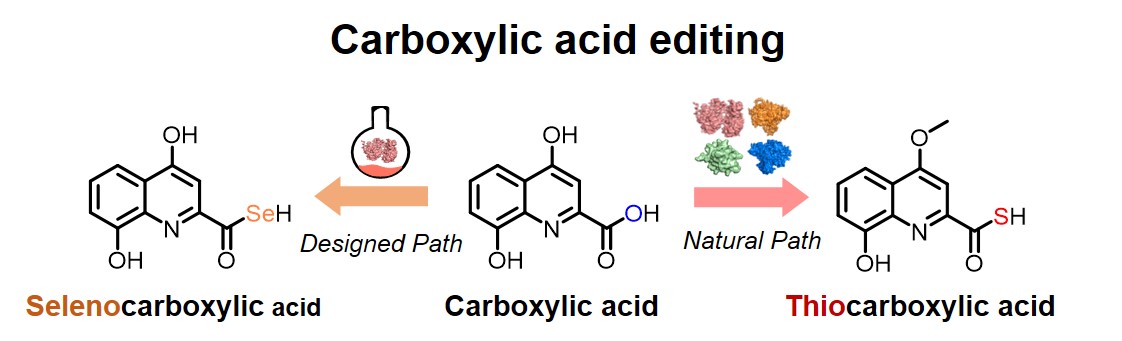
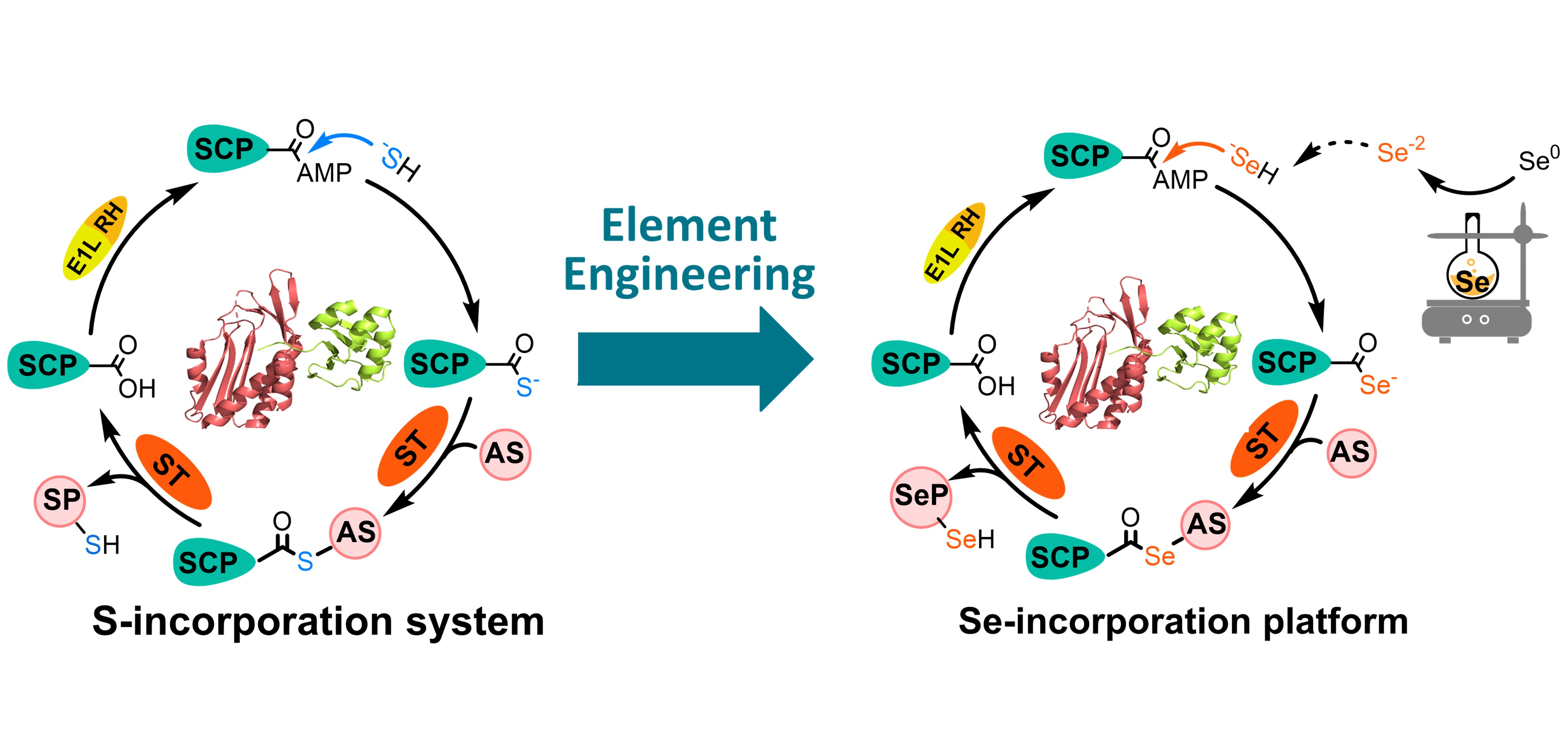
120. Zhang, X., Cheng, F., Guo, J., Zheng, S., Wang, X., Li, S. * Enzymatic synthesis of organoselenium compounds via C‒Se bond formation mediated by sulfur carrier proteins. Nat. Synth. 2024, 3, 477-487. | PDF
119. Liu, M., Li, S. * Nitrile biosynthesis in nature: how and why? Nat. Prod. Rep. 2024, 41, 649-671. | PDF

118. Zhang, C., Zhang, G., Chen, Y., Zheng, S., Du, J., Zhao, Z., Zhao, Y., Wang, N., Chen, C., Gao, Z. *, Li, S. *, Liu, K. * Sphingobacterium tenebrionis sp. nov., isolated from intestine of mealworm. Int. J. Syst. Evol. Microbiol. 2024, 74, 006455. | PDF
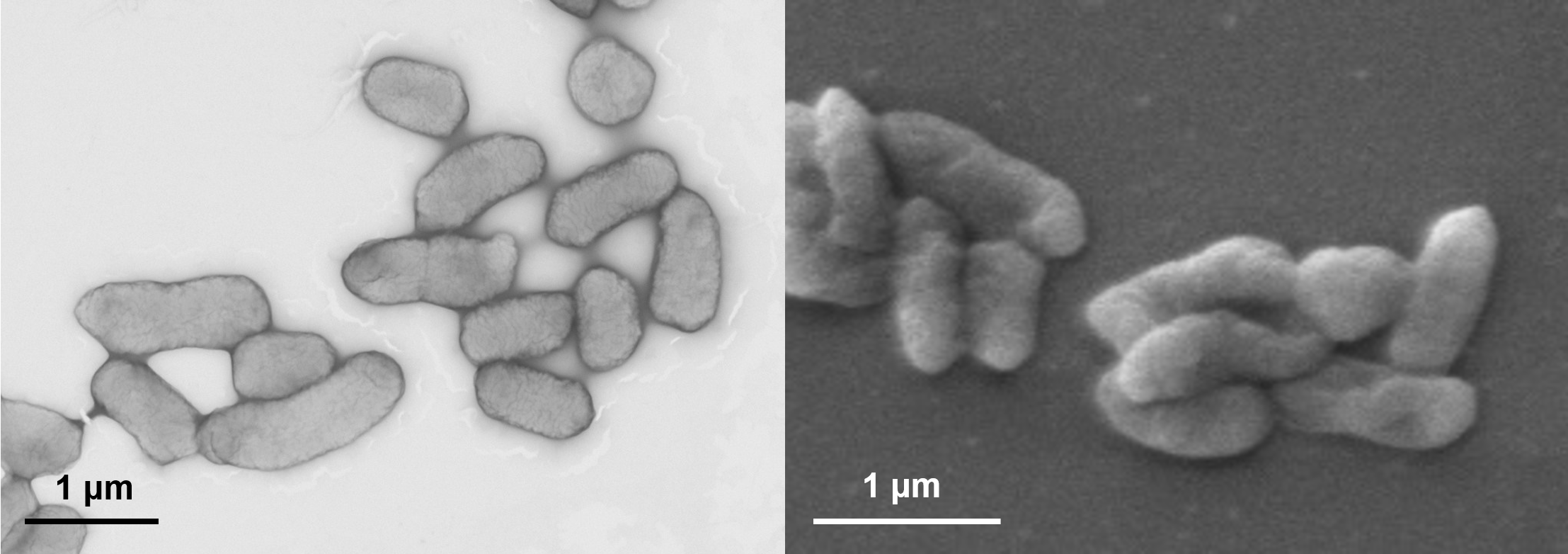
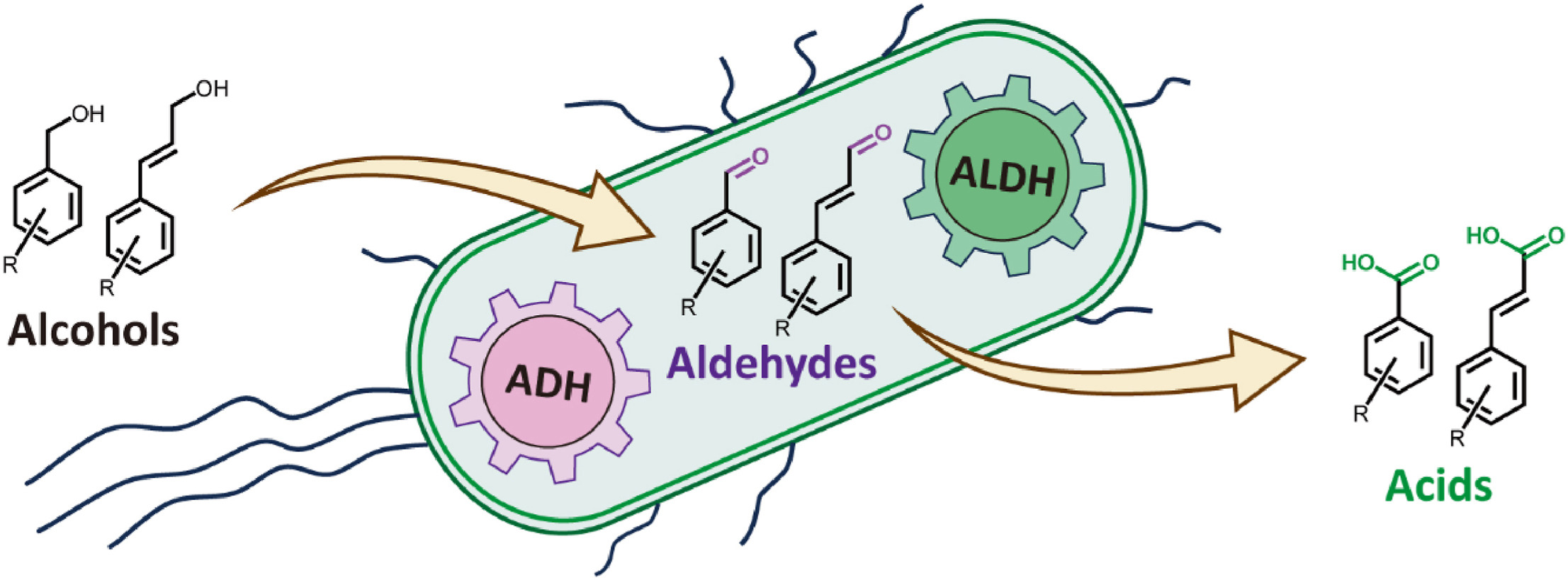
117. Qiu. Z. #, Liu, X. #, Yu, J., Zhao, Y., Zhao, G., Li, S., Liu, K. *, Du, L. *, Ma, L. * Efficient conversion of aromatic and phenylpropanoid alcohols to acids by the cascade biocatalysis of alcohol and aldehyde dehydrogenases. Synth. Syst. Biotechnol. 2024, 9(2), 187-195. | PDF
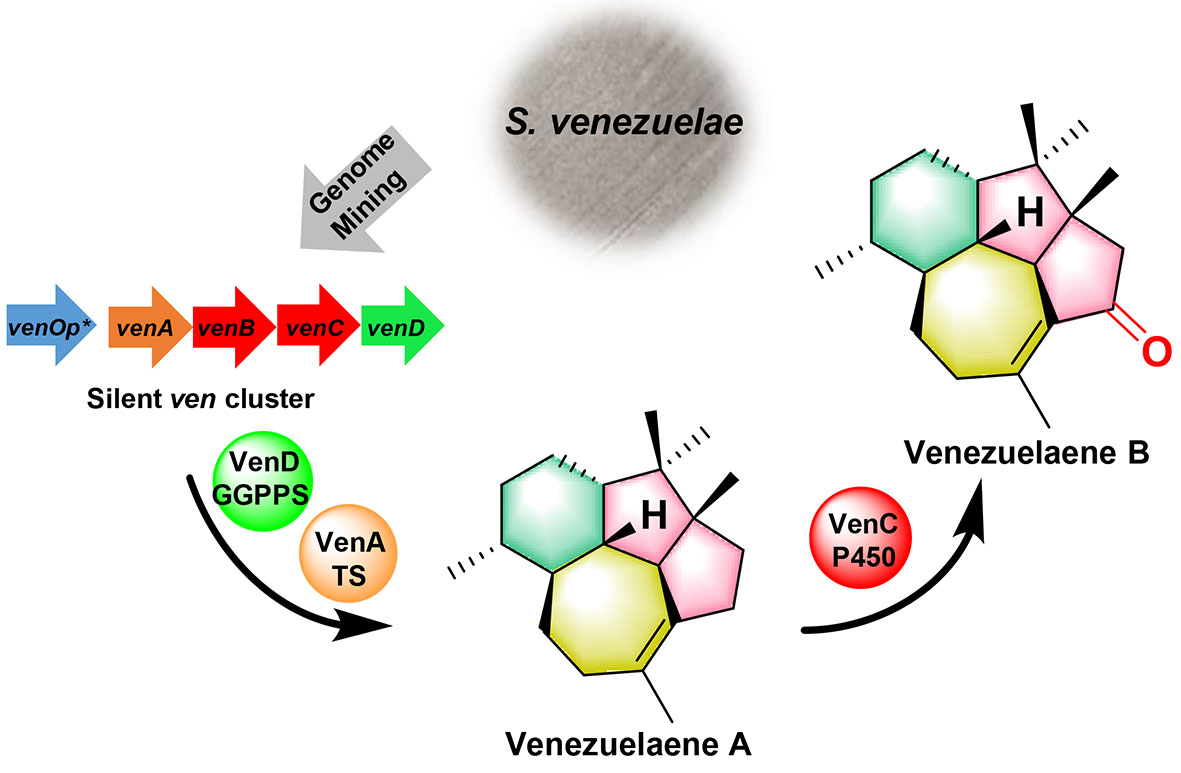
116. Li, S., Li, Z., Zhang, G., Urlacher. B. V., Ma, L. *, Li, S. * Functional analysis of the whole CYPome and Fdxome of Streptomyces venezuelae ATCC 15439. Eng. Microbiol. 2024, 4, 100166. | PDF

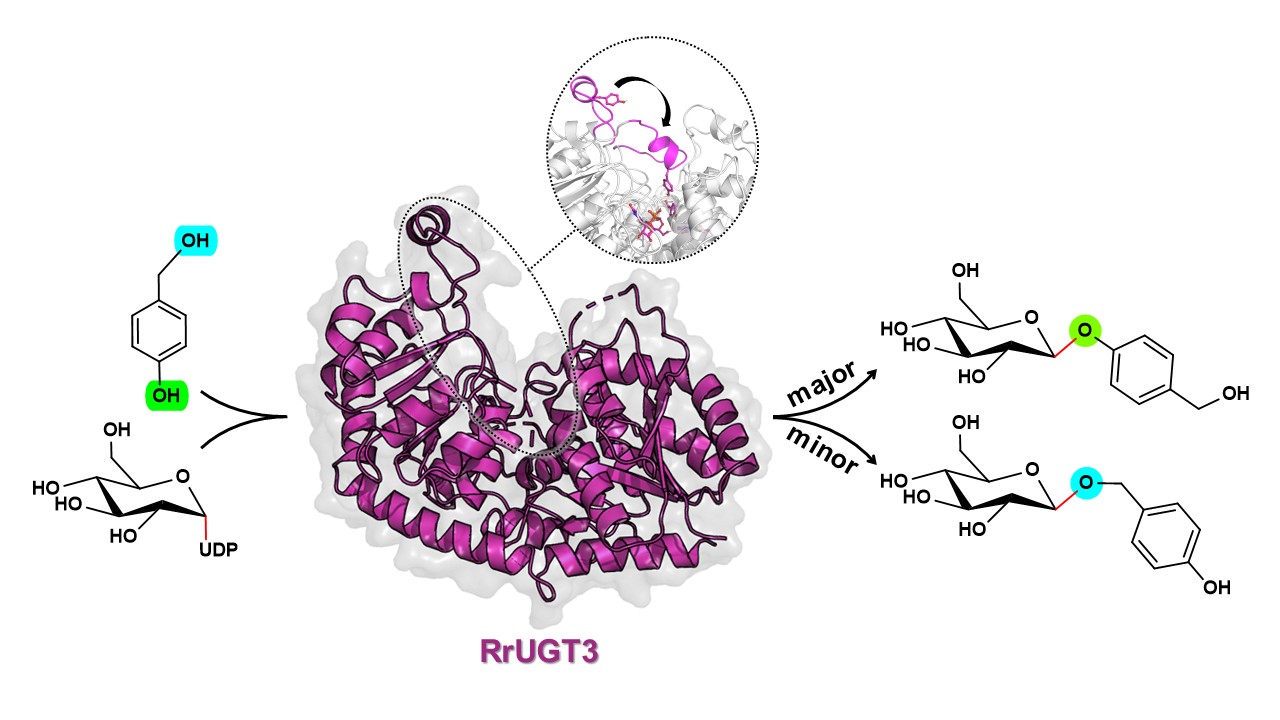
115. Li, M.#, You, C.#, Guo, F.#, Han, Q., Xie, X., Ma, L., Li, S., Du, L.*, Sheng, X.*, Su, H.* Reaction mechanism and regioselectivity of uridine diphosphate glucosyltransferase RrUGT3: a combined experimental and computational study. Catal. Sci. Technol. 2024, 14, 4882.| PDF
114. Sui, X., Wang, J., Zhao, Z., Liu, B., Liu, M., Liu, M., Shi, C., Feng, X., Fu, Y., Shi, D., Li, S., Qi, Q., Mian. M., Zhao, G. * Phenolic compounds induce ferroptosis-like death by promoting hydroxyl radical generation in the Fenton reaction. Commun. Biol. 2024, 7,199. | PDF
113. Zhu, L., Yang, Q., Wang, D., Niu, L., Pan, Z., Li, S., Li, Y., Zhang, W., Wu, C. Deciphering the Biosynthesis and Physiological Function of 5‑Methylated Pyrazinones Produced by Myxobacteria. ACS Cent. Sci. 2024, 10, 555-568. | PDF
112. Yang, D., Liu, H., Li, X., Zhang, Y., Zhang, X., Yang, H., Liu, M., Koch, E. K., McCarty, R. D., Li, S., Tan, B. * A sucrose ferulate cycle linchpin for feruloylation of arabinoxylans in plant commelinids. Nat. Plants 2024, 10, pages1389-1399. | PDF
2023
111. Xia, Y. #, Zhu, G. #, Zhang, X. #, Li, S., Du, L.*, Zhu, W.* Biosynthesis of 4‑Acyl-5-aminoimidazole Alkaloids Featuring a New Friedel−Crafts Acyltransferase. J. Am. Chem. Soc. 2023, 145: 26308-26317. | PDF
110. Liu, K., Gao, W., Xu, W., Liu, H.* Deep-sea microorganisms acquired during Jiaolong expedition. The Innovation Life 2023, 1(2): 100029. | PDF

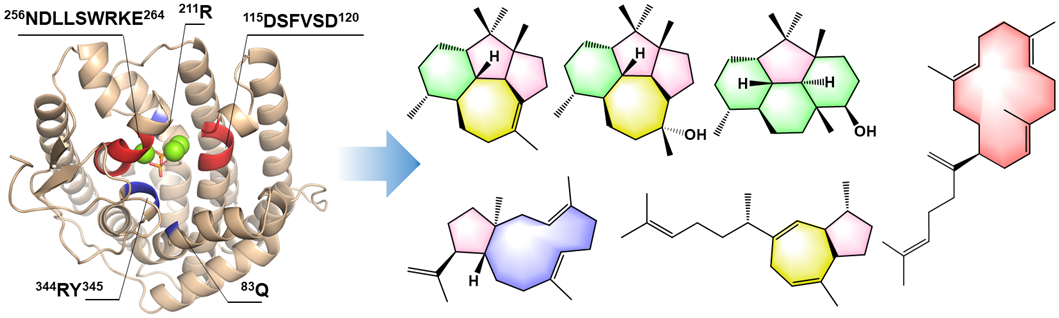
109. Li, Z. #, Zhang, L. #, Kang, X. #, Jiang, Y., Du, J., Zhang, X., Meng, L., Wu, Q., Du, L., Li, X., Hu, Y., Xie, Z., Jiang, X., Tang, Y., Wu, R. *, Guo, R. *, Li, S. * Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase. Nat. Commun. 2023, 14: 4001. | PDF
108. Ma, L., Sun, T., Liu, Y., Zhao, Y., Liu, X., Li, Y., Chen, X., Cao, L., Kang, Q., Guo, J., Du, L., Wang, W. *, Li, S. * Enzymatic synthesis of indigo derivatives by tuning P450 BM3 peroxygenases. Synth. Syst. Biotechnol. 2023, 8: 452-461. | PDF
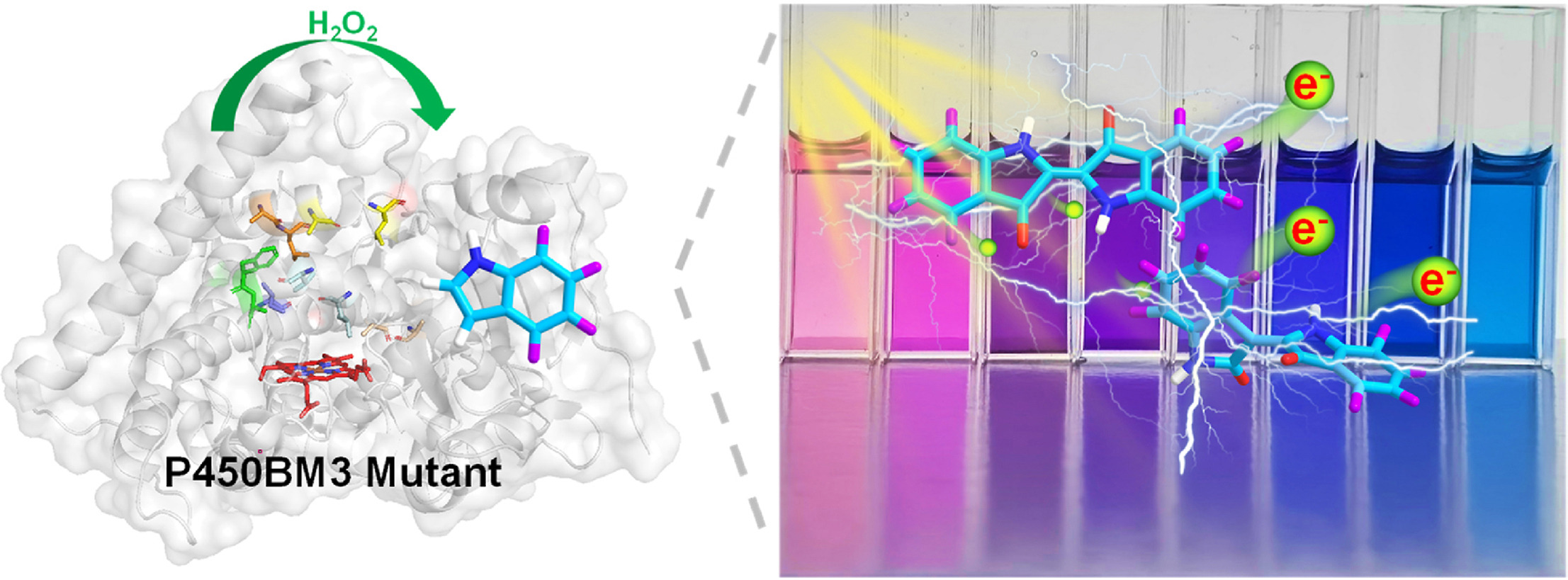
107. Pan, Y.#, Li, G.#, Liu, R., Guo, J., Liu, Y., Liu, M., Zhang, X., Chi, L., Xu, K., Wu, R., Zhang, Y., Li, Y., Gao, X.*, Li, S.* Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids. Nat. Commun. 2023, 14:1669. | PDF
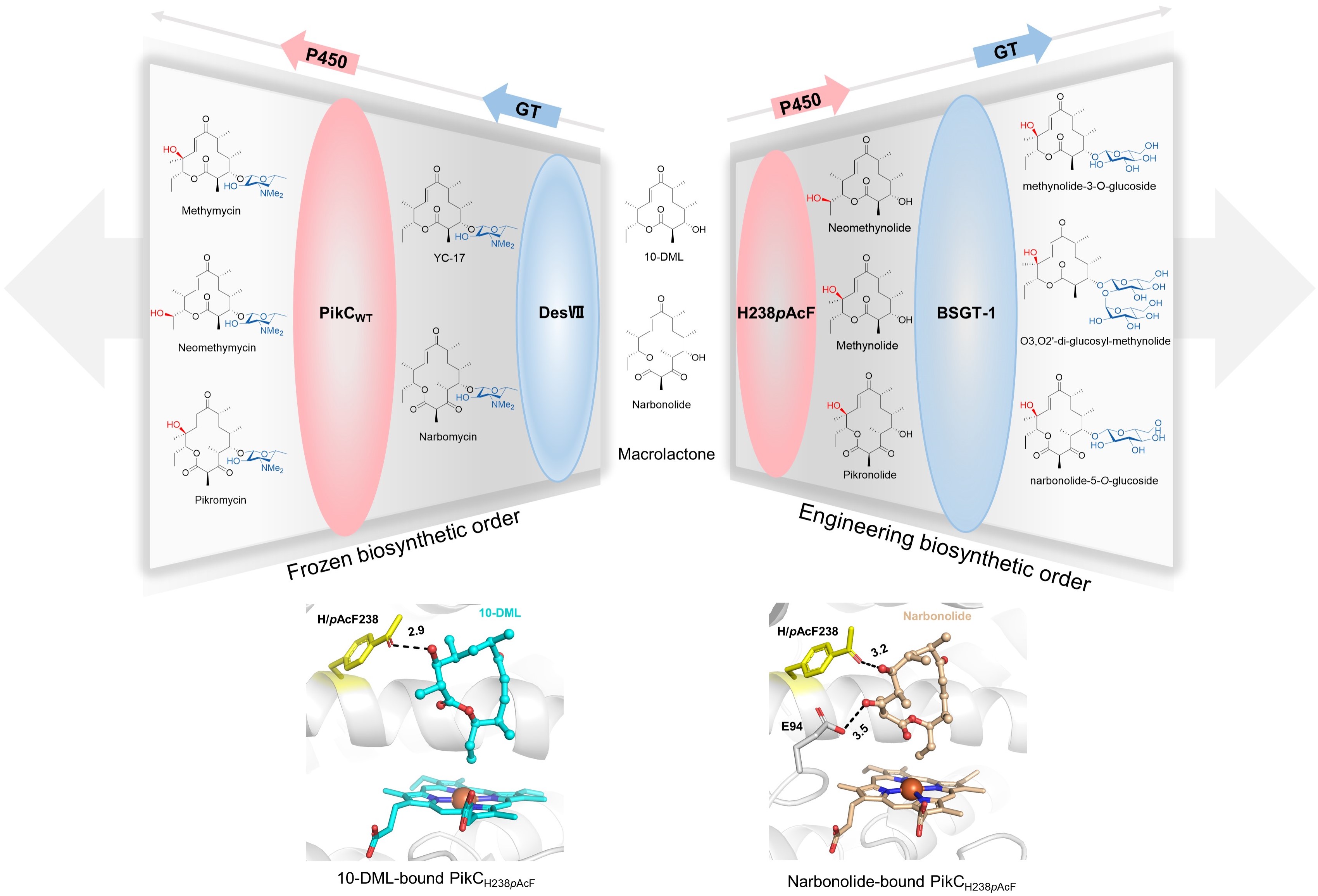
106. Liu, R., Pan, Y., Wang, N., Tang, D., Urlacher, B. V., Li, S.* Comparative biochemical characterization of mammalian-derived CYP11A1s with cholesterol side-chain cleavage activities. J. Steroid Biochem. 2023, 229, 106268. | PDF
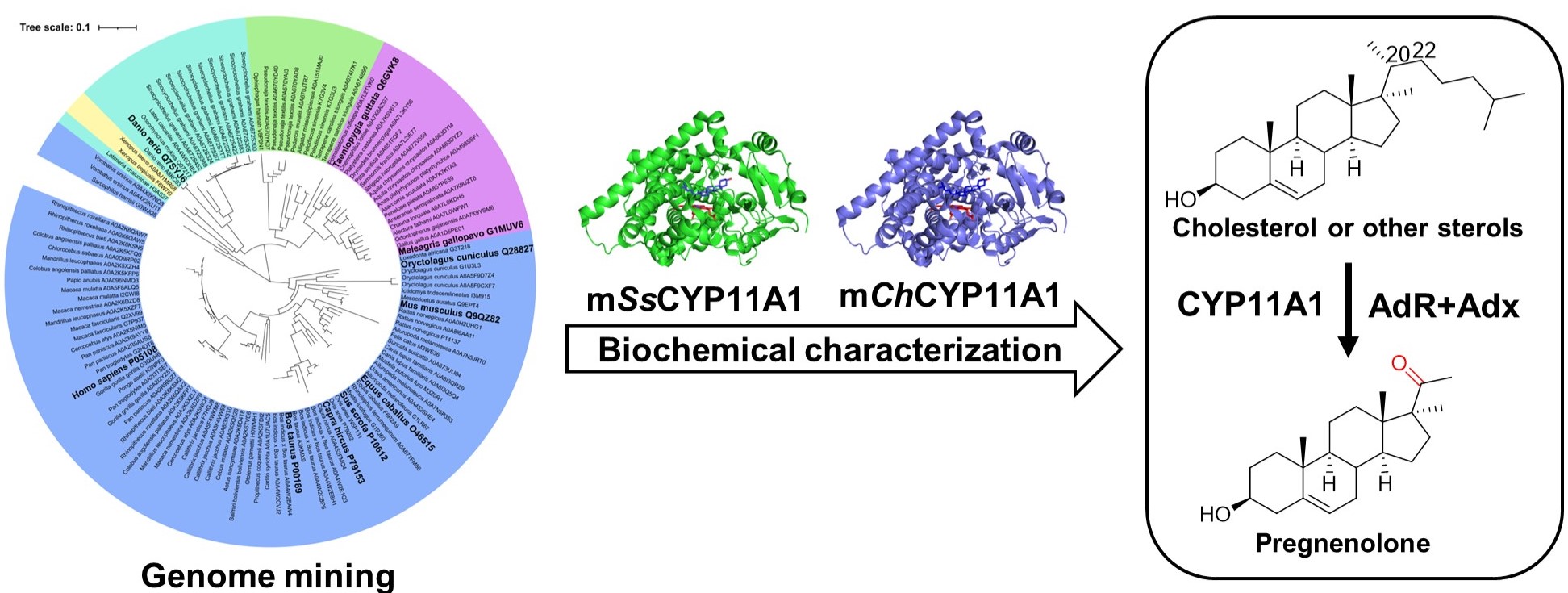
105. Yang, F.#, Sang, M.#, Liu, J.-R., Zhao, H.-M.,Zou, Y., Wu, W., Yu, Y., Ma, W., Zhang, Y., Zhang, W.*, Lin, H.-W.* Somalactams A–D: Anti-inflammatory Macrolide Lactams with Unique Ring Systems from an Arctic Actinomycete Strain. Angew. Chem. Intl. Ed. 2023, 62, e202218085. | PDF
104. Chi, X. *, Zhao, Z., Han, Q., Yan, H., Ji, B., Chai, Y., Li, S., Liu, K. * Insights into autotrophic carbon fixation strategies through metagonomics in the sediments of seagrass beds. Mar. Environ. Res. 2023, 188, 106002. | PDF
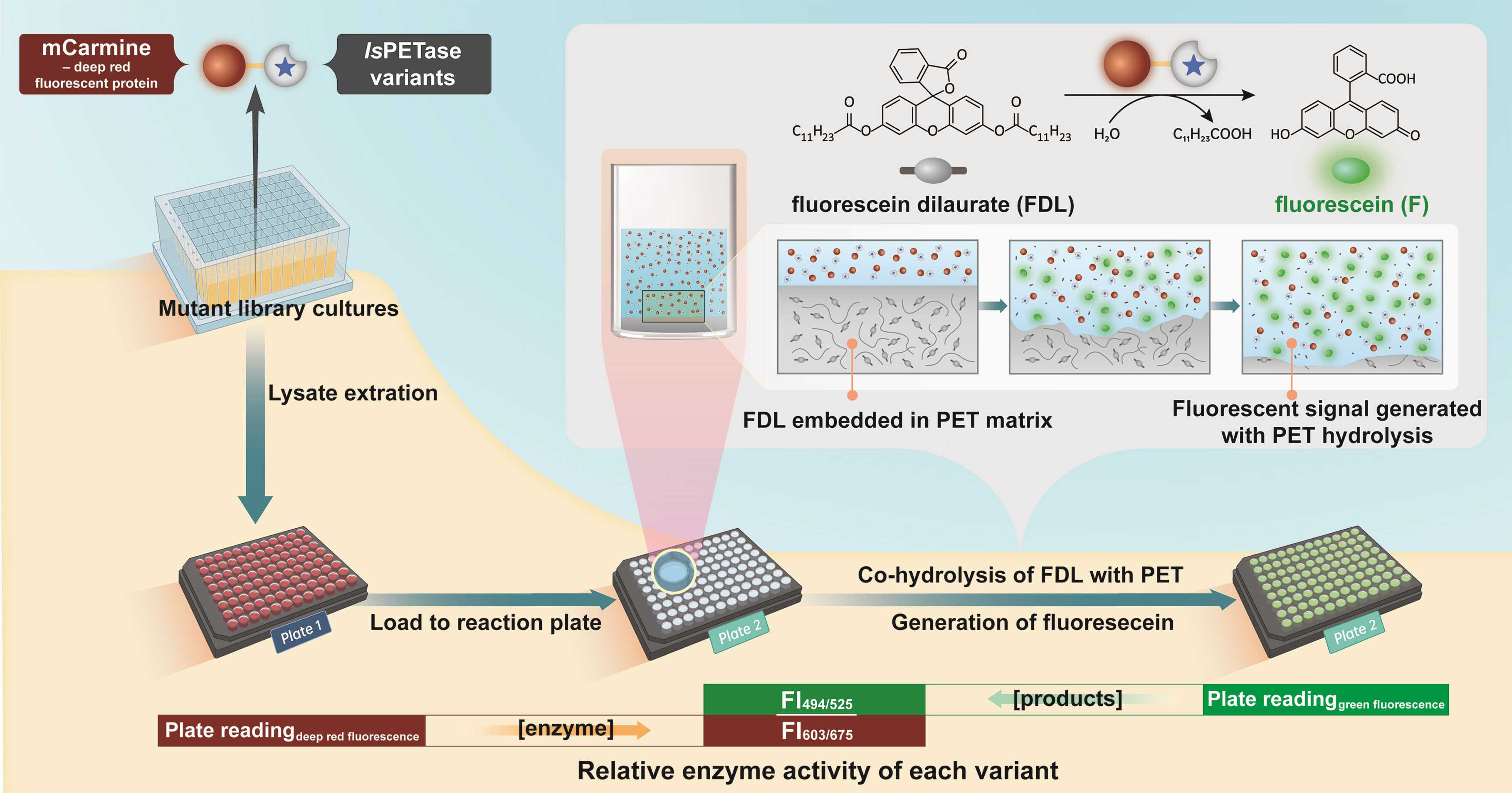
103. Liu, K., Xu, Z., Zhao, Z., Chen, Y., Chai, Y., Ma, L., Li, S.* A dual fluorescence assay enables high-throughput screening for PET hydrolases. ChemSusChem 2023, 16, e2022020. | PDF
102. Luelf, U. J.,Böhmer, M. L., Li, S., Urlacher, B. V. * Effect of chromosomal integration on catalytic performance of a multi‐component P450 system in Escherichia coli. Biotechnol. Bioeng. 2023, 120(7), 1762-1772. | PDF
2022
101. Li, S. #, Chi, L. #, Li, Z., Liu, M., Liu, R., Sang, M., Zheng, X., Du, L., Zhang, W., Li, S.* Discovery of venediols by activation of a silent type I polyketide biosynthetic gene cluster in Streptomyces venezuelae ATCC 15439. Tetrahedron 2022, 133072. | PDF

100. Guo, F. #, Zhang, X. #, You, C., Zhang, C., Li, F., Li, N., Xia, Y., Liu, M., Qiu, Z., Zheng, X., Ma, L., Zhang, G., Luo, L., Cao, F., Feng, Y., Zhao, G., Zhang, W., Li S., Du, L.* Diversifcation of phenolic glucosides by two UDP-glucosyltransferases featuring complementary regioselectivity. Micro. Cell. Fact. 2022, 21, 208. | PDF
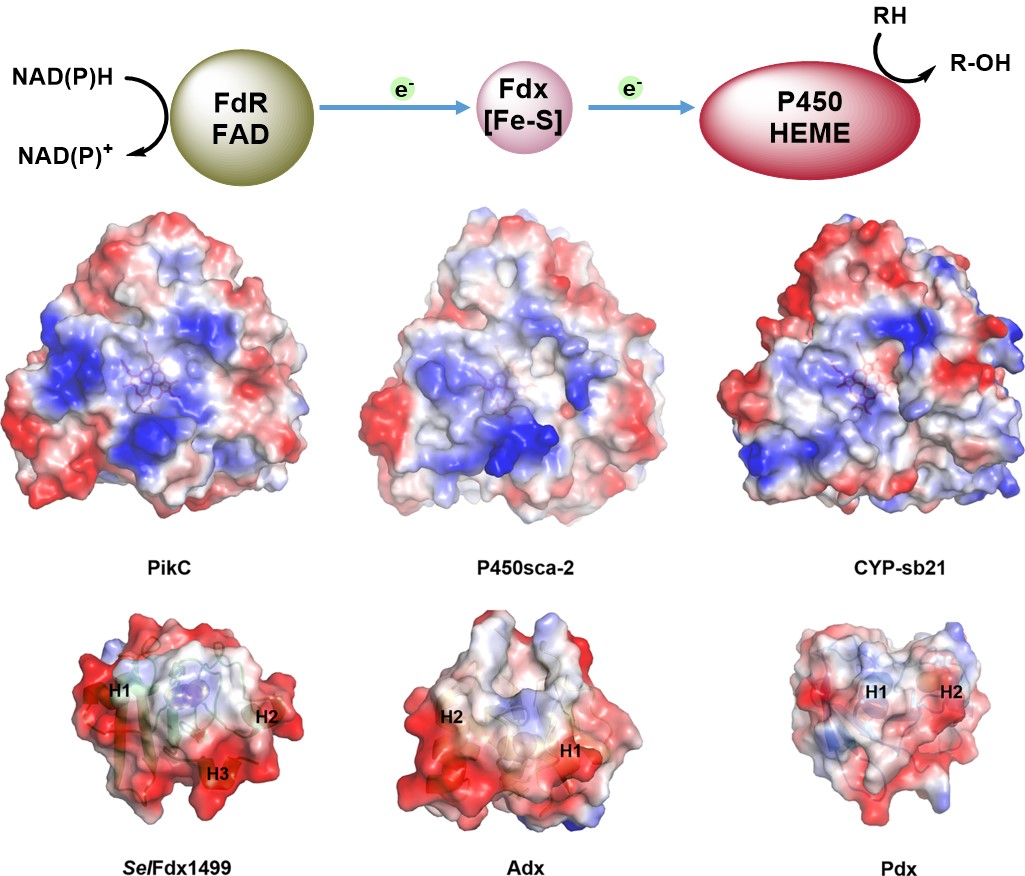
99. Liu, X.#, Li, F.#, Sun, T., Guo, J., Zhang, X., Zheng, X., Du, L., Zhang, W., Ma, L. *, Li, S. Three pairs of surrogate redox partners comparison for Class I cytochrome P450 enzyme activity reconstitution. Commun. Biol 2022, 5(1), 791. | PDF
98. Zheng, S. #, Guo, J. #, Cheng, F., Gao, Z., Du, L., Meng, C.*, Li, S.*, Zhang, X.* Cytochrome P450s in algae: Bioactive natural product biosynthesis and light-driven bioproduction. Acta Pharm. Sin. B 2022,12(6), 2832-2844. | PDF
96. Wang, Y. #, Wang, P. #, Cao, H., Ding, H., Su, H., Liu, S., Liu, G., Zhang, X., Li, C., Peng, M., Li, F., Li, S., Chen, Y., Chen, X. *, Zhang, Y. * Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis. Nat. Commun. 2022, 13(1): 566. | PDF
95. Gao, X. #, Su, Q. #, Li, J., Yang, W., Yao, B., Guo, J., Li, S., Liu C. * RNA-Seq analysis reveals the important co-expressed genes associated with polyphyllin biosynthesis during the developmental stages of Paris polyphylla. BMC Genom. 2022, 23, 559. | PDF
94. Wu, M. #, Miao, L. #, Liu, Y. #, Qian, X., Hou, T., Ai, G., Yu, L., Ma, L., Gao, X., Qin, Y., Zhu, H., Du, L., Li, S., Tian, C., Li, D. *, Liu, Z. *, Liu, S. * Identification and characterization of a novel hydroxylamine oxidase, DnfA, that catalyzes the oxidation of hydroxylamine to N2. J. Biol. Chem. 2022, 298(9), 102372. | PDF
2021
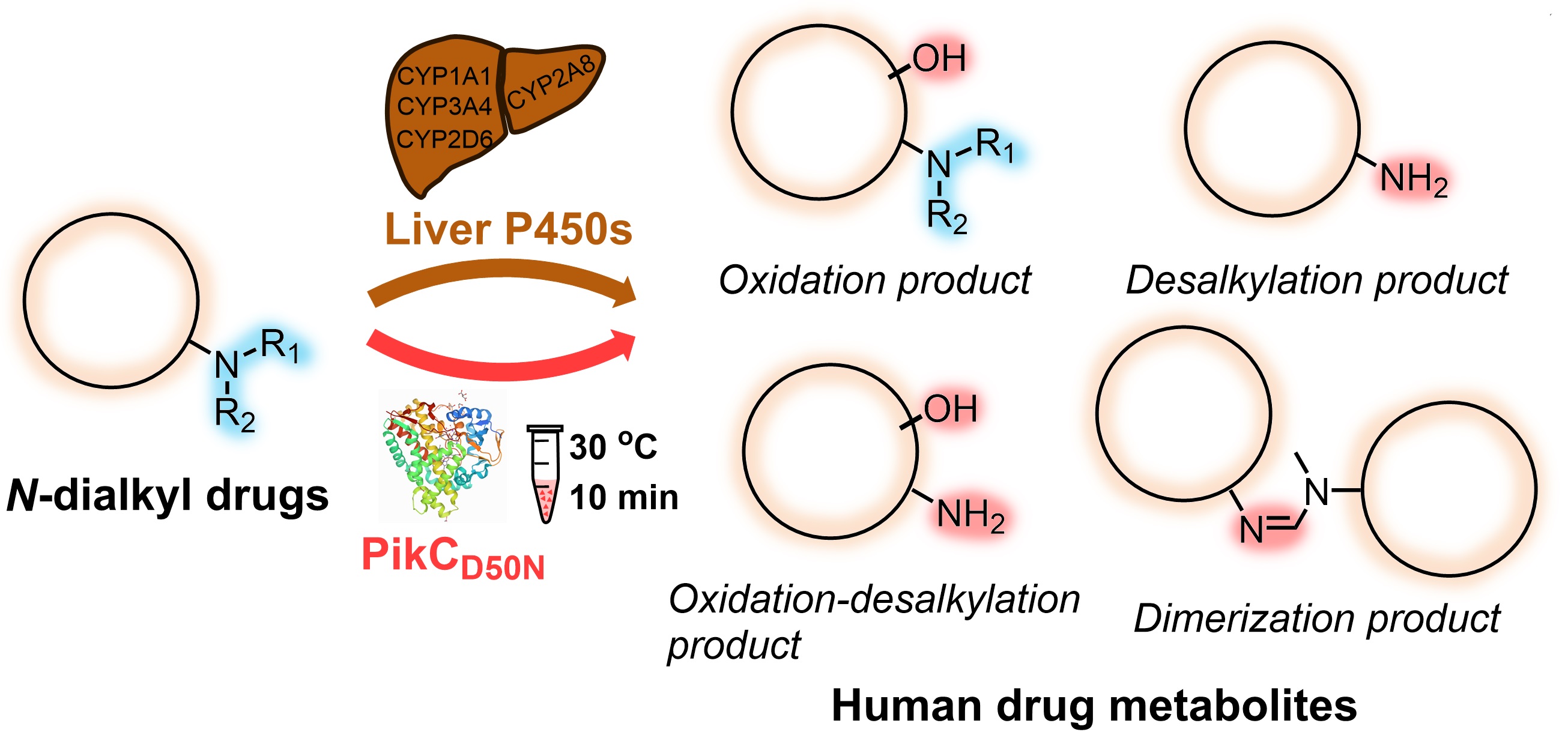
93. Guo, J., Li, F., Cheng, F., Ma, L., Liu, X., Durairaj, P., Zhang, G., Tang, D., Long, X., Zhang, W., Du, L., Zhang, X.*, Li, S. Bacterial biosynthetic P450 enzyme PikCD50N: A potential biocatalyst for the preparation of human drug metabolites.J. Org. Chem.2021,86, 14563-14571.|PDF
92. Jiang, Y. #, Peng, W. #, Li, Z. #, You, C., Zhao, Y., Tang, D., Wang, B. *, Li, S.*Unexpected reactions of α,β-unsaturated fatty acids provide insight into the mechanisms of CYP152 peroxygenases.Angew. Chem. Intl. Ed.2021, 60, 24694-24701.|PDF
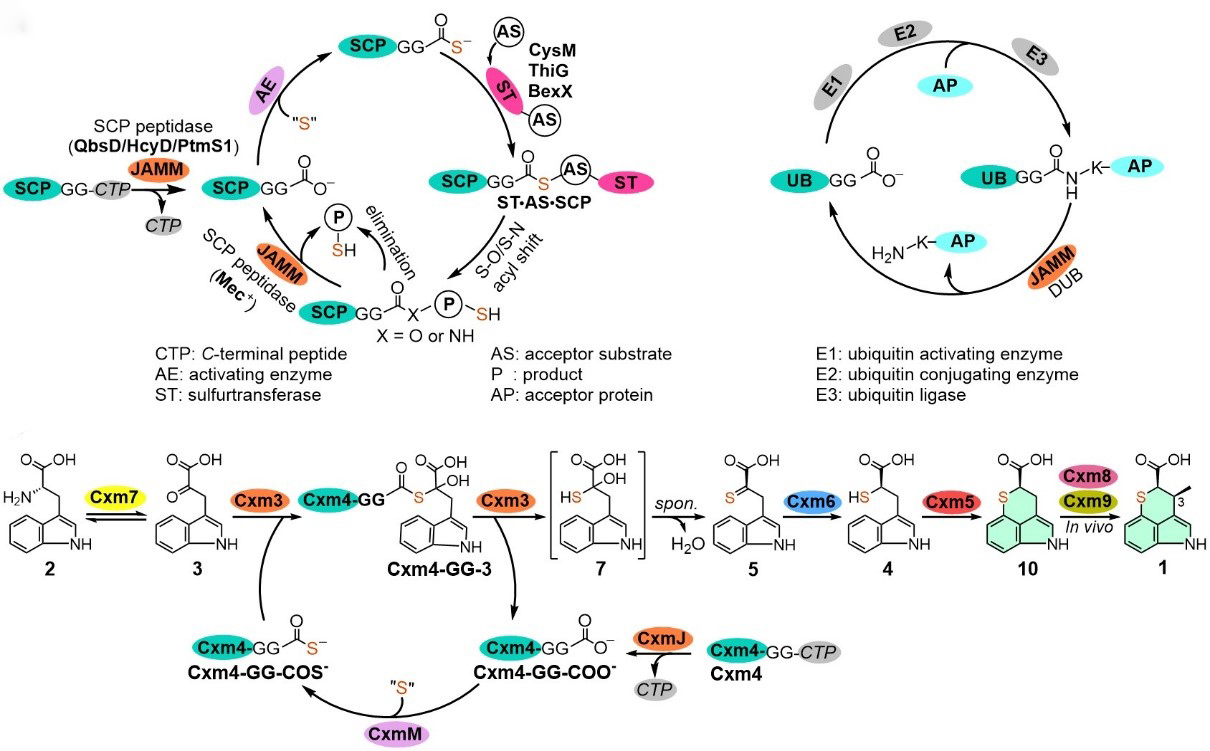
91. Zhang, X., Xu, X., You, C., Yang, C., Guo, J., Sang, M., Geng, C., Cheng, F., Du, L., Shen, Y., Wang, S., Lan, H., Yang, F., Li, Y., Tang, Y., Zhang, Y., Bian, X.*, Li, S.*, Zhang, W.* Biosynthesis of chuangxinmycin featuring a deubiquitinase-like sulfurtransferase.Angew. Chem. Intl. Ed.2021, 133, 24623-24628.|PDF
90. Ma, L., Li, F., Zhang, X., Chen, H., Huang, Q., Su, J., Liu, X., Sun, T., Fang, B., Liu, K., Tang, D., Wu, D., Zhang, W., Du, L., Li, S. * Development of MEMS directed evolution strategy for multiplied throughput and convergent evolution of cytochrome P450 enzymes.Sci. China Life Sci.2021, 65(3), 550-560.|PDF

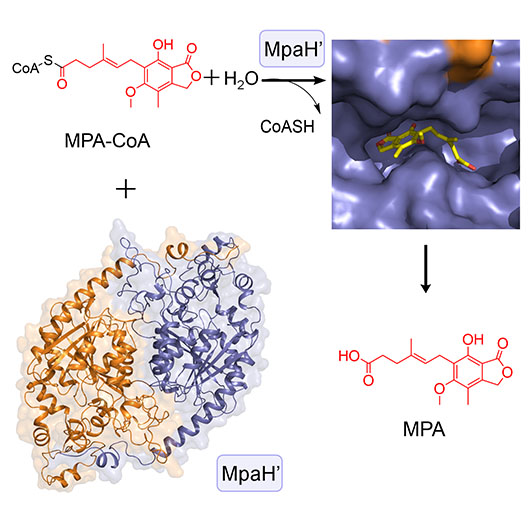
89. You, C., Li, F., Zhang, X., Ma, L., Zhang, Y., Zhang, W., and Li, S. * Structural basis for substrate speci?city of the peroxisomal acyl-CoA hydrolase MpaH’involved in mycophenolic acid biosynthesis.FEBS J.2021, 288, 5768-5780.|PDF
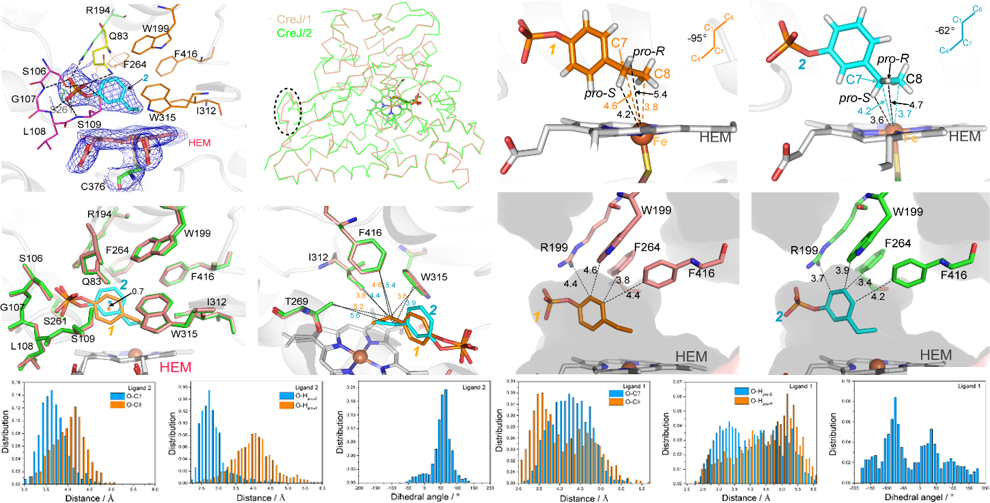
88. Dong, S. #, Chen, J. #, Zhang, X., Guo, F., Ma, L., You, C., Wang, X., Zhang, W., Wan, X., Liu, S., Yao, L., Li, S., Du, L. * and Feng, Y. * Structural basis for selective oxidation of phosphorylated ethylphenols by cytochrome P450 monooxygenase CreJ.Appl. Environ. Microbiol.2021, 87(11), e00018-21.|PDF
87. Zhang, X., Guo, J., Cheng, F., and Li, S. * Cytochrome P450 enzymes in fungal natural product biosynthesis.Nat. Prod. Rep.2021, 38, 1072-1099.|PDF
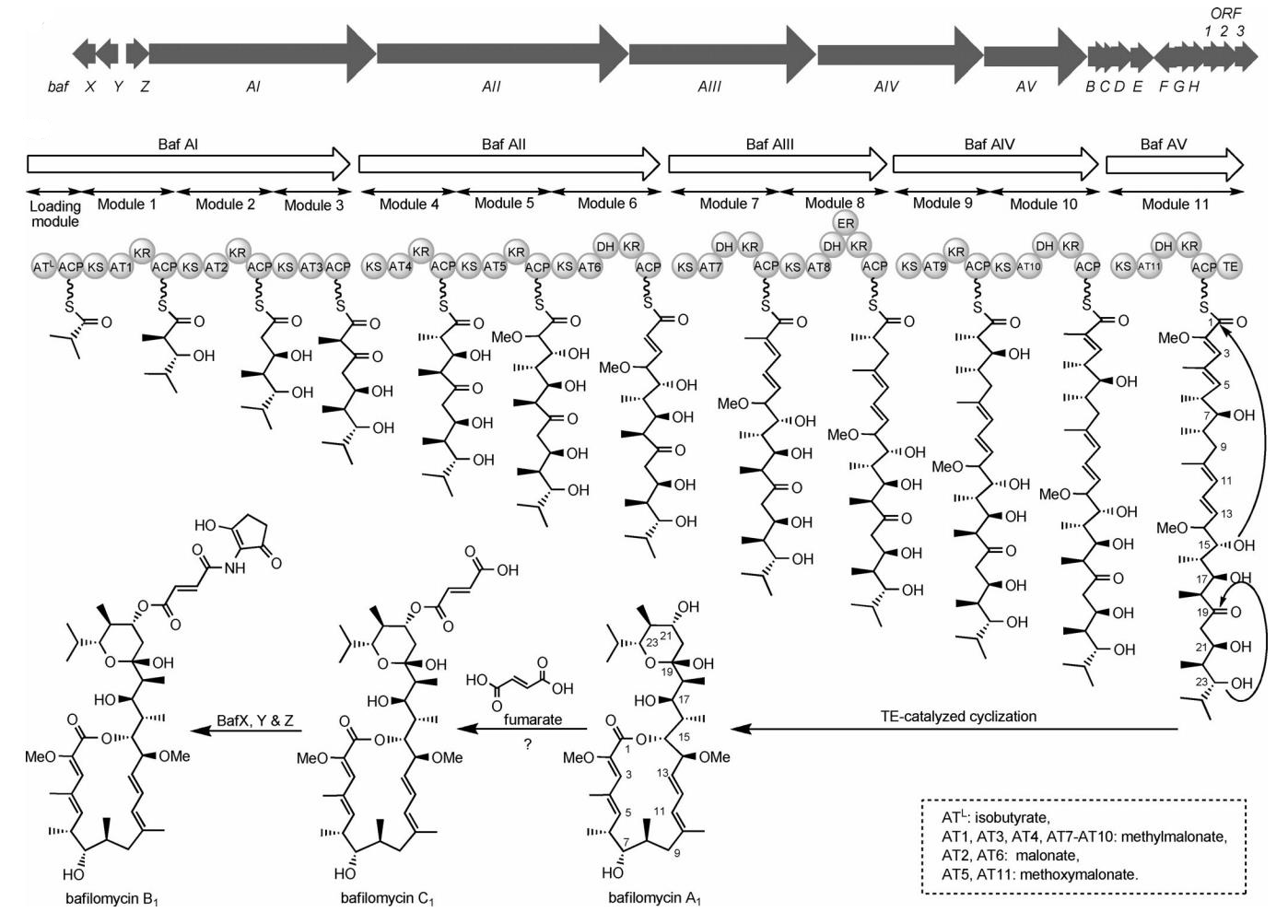
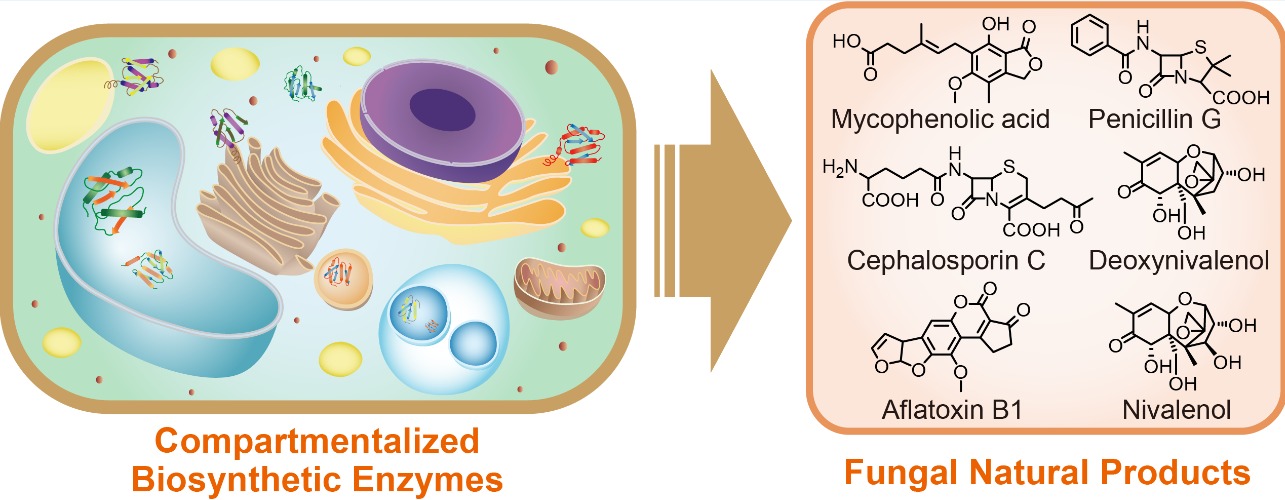
86. Du, L., and Li, S. * Compartmentalized biosynthesis of fungal natural products.Curr. Opin. Biotechnol.2021, 69, 128-135.|PDF
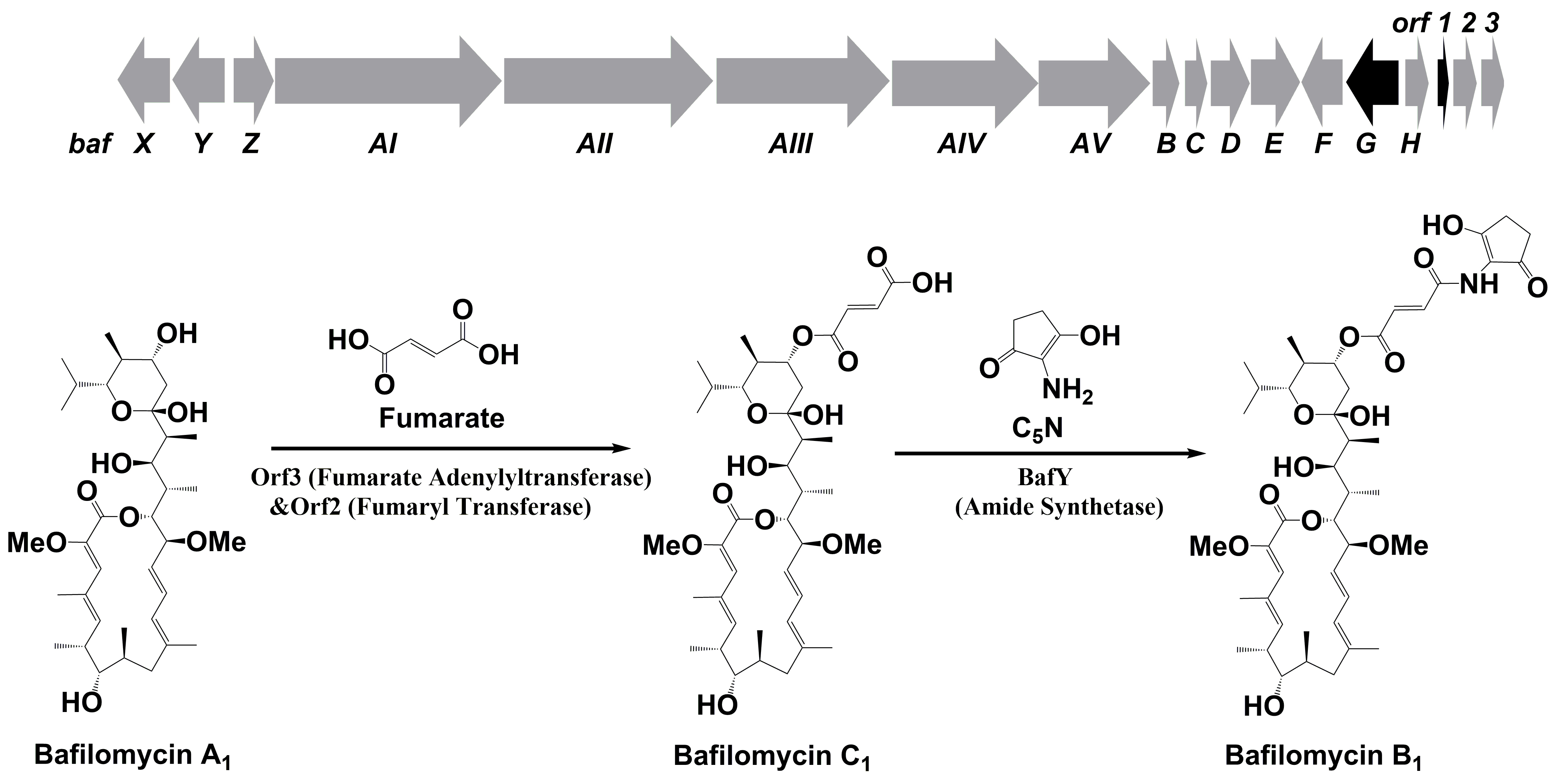
85. Li, Z., Li, S., Du, L., Zhang, X., Jiang, Y., Liu, W., Zhang, W., and Li, S. * Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine BacteriumStreptomyces lohii.Mar. Drugs, 2021, 19, 29.|PDF
84. Zhao, W. #, Duan, Y. #, Li, H., Li, S., Shen, Y., Zhang, Y., Li, Y., Tang, Y. * Triazole/thiadiazole substituted 4'-demethylepipodophyllotoxin derivatives induced apoptosis in HeLa cells by up-regulating TMEM133.Eur. J. Pharmacol.2021, 905, 174189.|PDF
83. Li, C. #, Wang, X. #, Chen, X., Sheng, Q., Zhang, S., Wang, P., Quareshy M., Rihtman, B., Shao, X., Gao, C., Li, F., Li, S., Zhang, W., Zhang, X., Yang, G., Todd, J. D., Chen, Y., Zhang, Y. * A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.elife2021, 10, e64045.|PDF
82. Cheng, Y., Zhu, S., Guo, C., Xie, F., Jung, D., Li, S., Zhang, W., * He, S. *Microbulbifer hainanensissp. nov., a moderately halopilic bacterium isolated from mangrove sediment.Antonie van Leeuwenhoek2021, 114(7), 1033-1042.|PDF
81. Xie, B. #, Rong, J. #, Tang, B. #, Wang, S. #, Liu, G., Qin, Q., Zhang, X., Zhang, W., She, Q., Chen, Y., Li, F., Li, S., Chen, X., Luo, H. *, Zhang, Y. * Evolutionary Trajectory of the Replication Mode of BacterialReplicons.mBio2021, 12(1), e02745-20.|PDF
2020
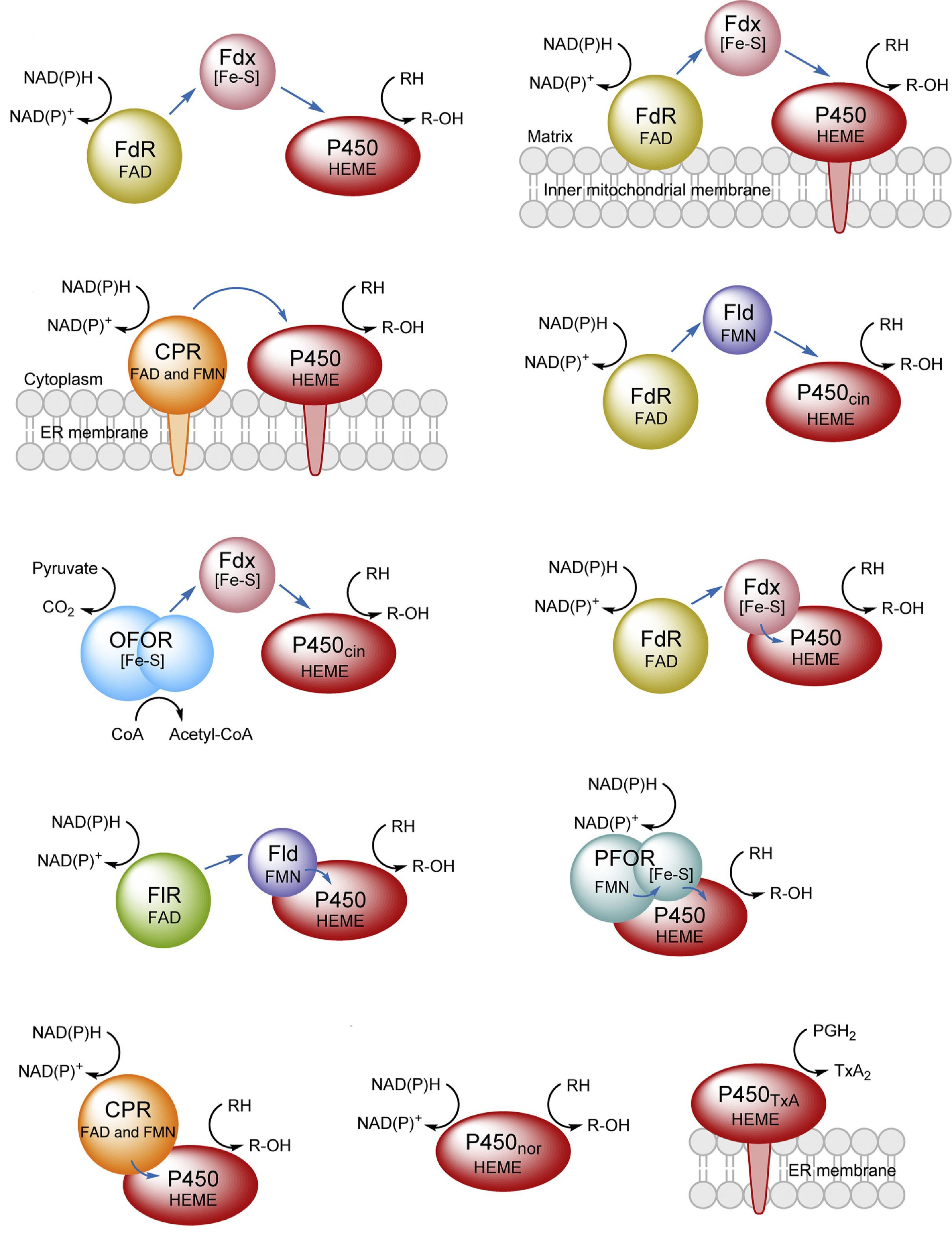
78. Li, S.*, Du, L., and Bernhardt, R.* Redox partners: Function modulators of bacterial P450 enzymes. Trends Microbiol. 2020, 28(6): 445-454. | PDF
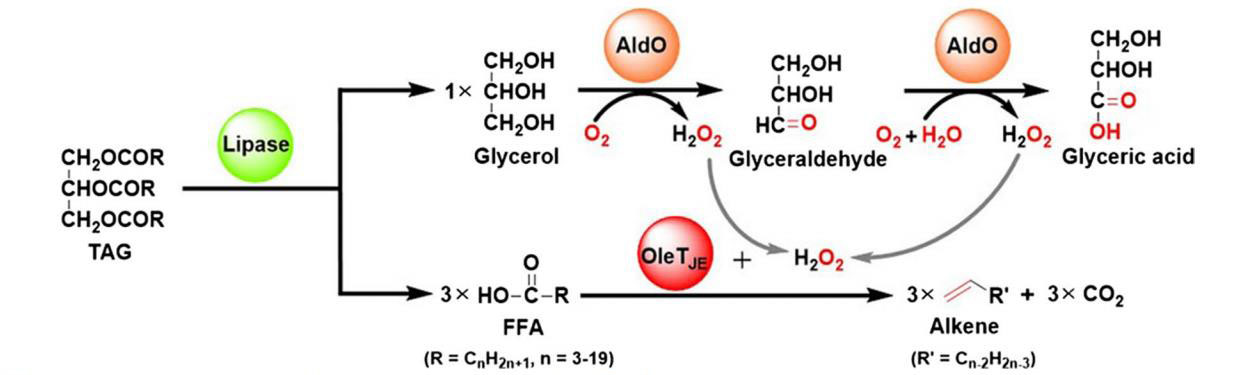
76. Li, Z. #, Jiang, Y. #, Guengerich, F. P., Ma, L., Li, S., and Zhang, W. * Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295(3), 833-849. | PDF
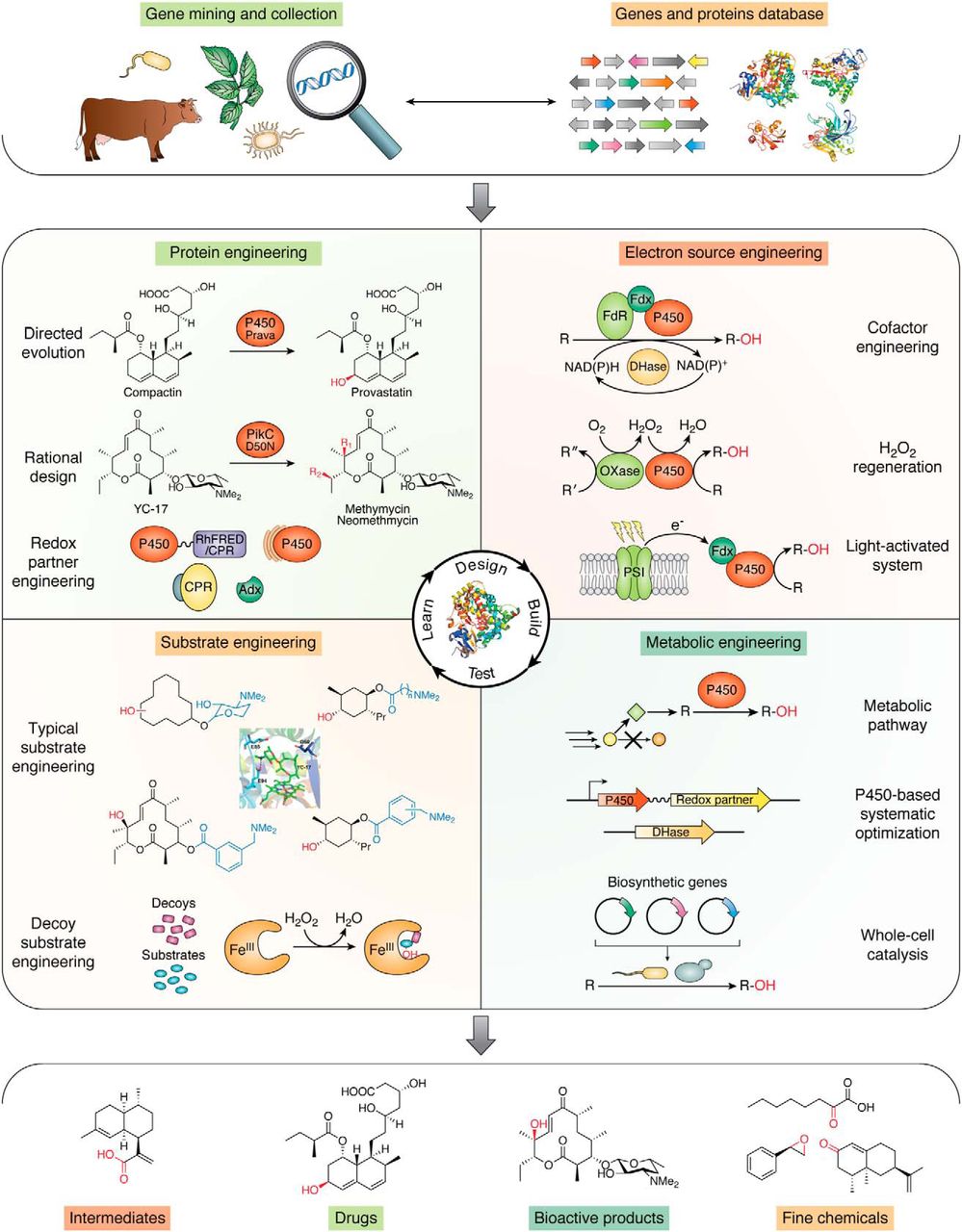
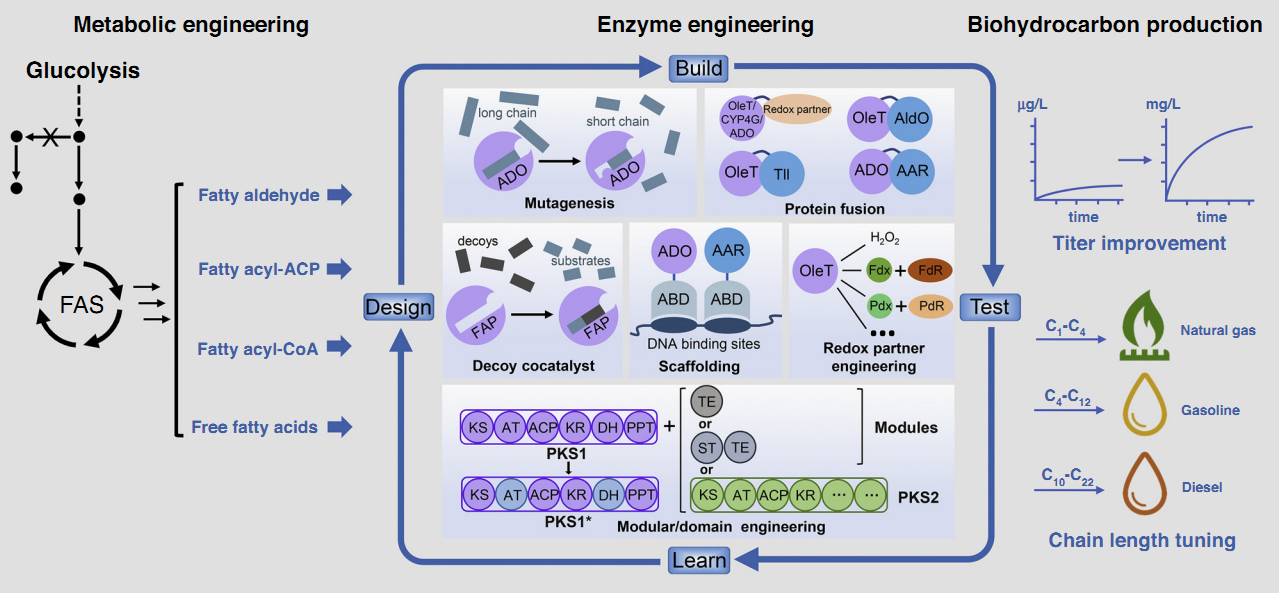
73. Li, F.*, Ma, L., Zhang, X., Chen, J., Qi, F., Huang, Y., Qu, Z., Yao, L., Zhang, W., Kim, E.-S.*, and Li, S.* Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana. Synth. Syst. Biotechnol. 2020, 5, 236-243. | PDF
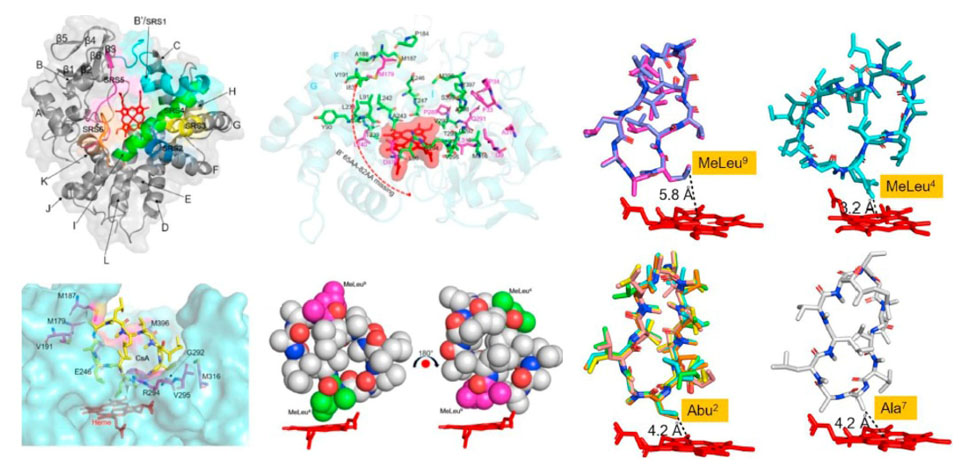
72. Yang, S., Cao, X., Yu, W., Li, S., Zhou, Y. J. * Efficient targeted mutation of genomic essential genes in yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2020, 104(7):3037-3047. | PDF
71. Ding, L., Xu, P., Zhang, W., Yuan, Y., He, X., Su, D., Shi, Y., Naman, C. B., Yan, X., Wu, B., Lazaro, J. E. H., Li, S., and He, S.* Three new diketopiperazines from the previously uncultivable marine bacterium Gallaecimonas mangrovi HK-28 cultivated by iChip. Chem. Biodivers. 2020,17, e2000221. | PDF
70. Fraley, A. E., Tran, H. T., Kelly, S. P., Newmister, S. A., Tripathi, A., Kato, H., Tsukamoto, S., Du, L., Li, S., Williams, R. M.*, and Sherman, D. H.* Flavin-dependent monooxygenases NotI and NotI’ mediate spiro-oxindole formation in biosynthesis of the notoamides. Chembiochem 2020, 21, 2449-2454. | PDF
69. Jia, K.#, Zhan, X.#, Li, H., Li, S., Shen, Y., Qi, Q., Zhang, Y., Li, Y., and Tang, Y.* A novel podophyllotoxin derivative with higher anti-tumor activity produced via 4?-demethylepipodophyllotoxin biotransformation by Penicillium purpurogenum. Process Biochem. 2020, 96, 220-227. | PDF
68. Gao, X., Zhang, X., Chen, W., Li, J., Yang, W., Zhang, X., Li, S., Liu, C.* Transcriptome analysis of Paris polyphylla var. yunnanensis illuminates the biosynthesis and accumulation of steroidal saponins in rhizomes and leaves. Phytochemistry 2020, 178, 112460.| PDF
67. Zhao, W. #, Cong, Y. #, Li, H., Li, S., Shen, Y., Qi, Q., Zhang, Y., Li, Y., Tang, Y. * Challenges and potential for improving the druggability of podophyllotoxin-derived drugs in cancer chemotherapy. Nat. Prod. Rep. 2020,38(3), 470-488.| PDF
2019
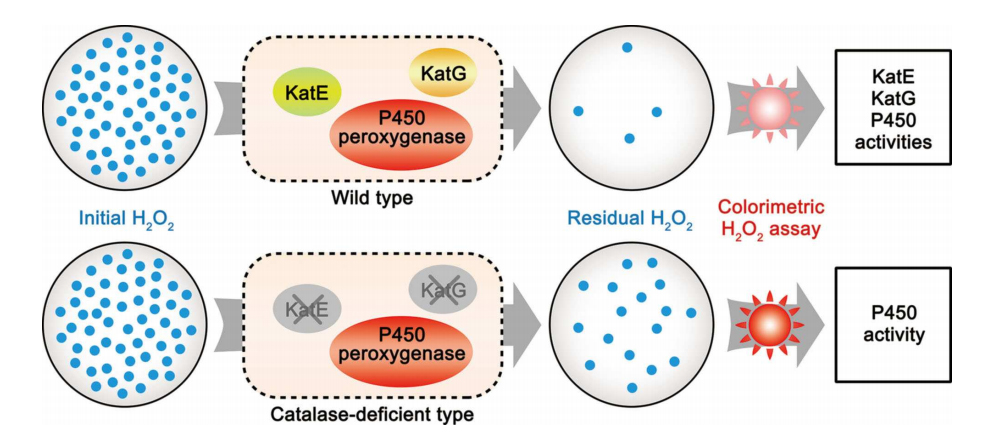
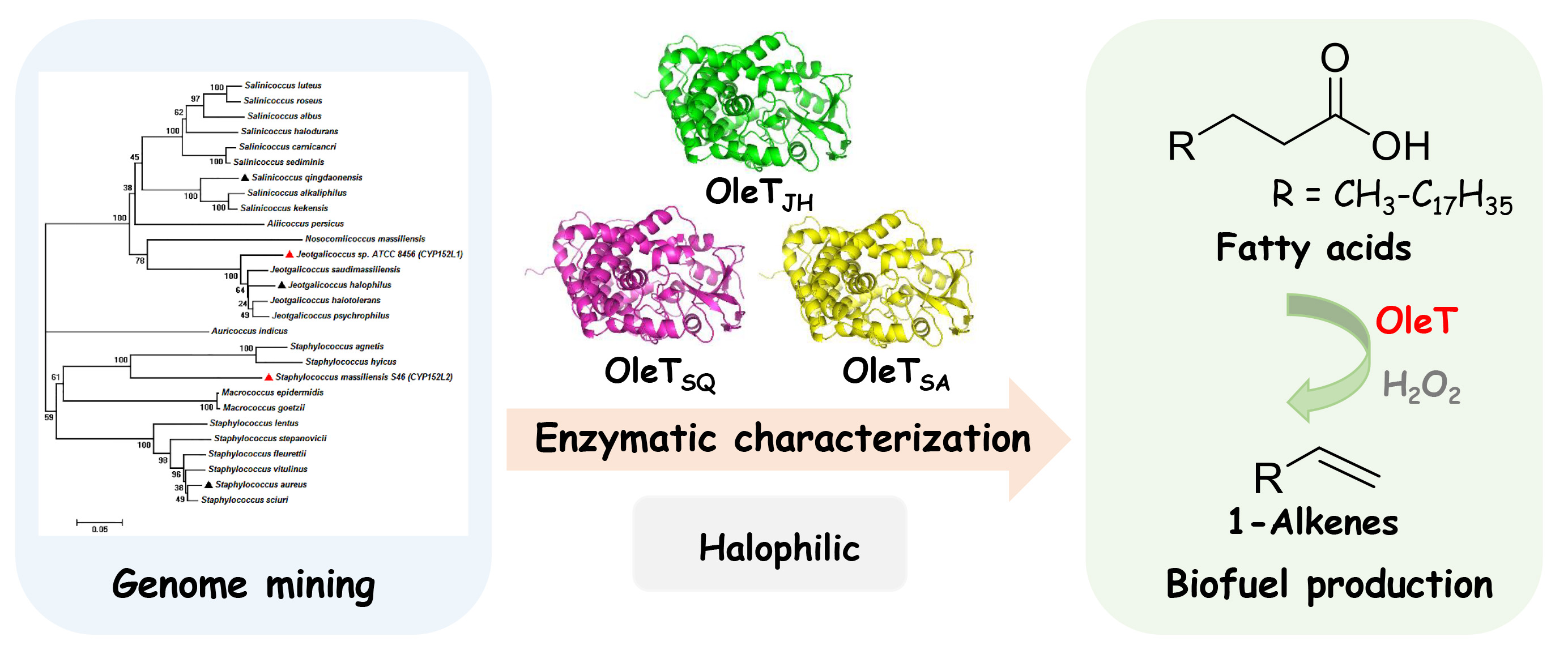
64. Jiang, Y., Li, Z., Wang, C., Zhou, Y., Xu, H.*, and Li, S.* Biochemical characterization of three new α-olefn-producing P450 fatty acid decarboxylases with a halophilic property. Biotechnol. Biofuels 2019, 12, 79. | PDF
63. Huang, Y., Zheng, X., Pilgaard, B., Holck, J., Muschiol, J., Li, S. and Lange, L. Identification and characterization of GH11 xylanase and GH43 xylosidase from the chytridiomycetous fungus, Rhizophlyctis rosea. Appl. Microbiol. Biotechnol. 2019, 103, 777-791. | PDF
62. Jia, K., Zhu, L., Qu, X., Li, S., Shen, Y., Qi, Q., Zhang, Y., Li, Y., and Tang, Y.* Enzymatic o-glycosylation of etoposide aglycone by exploration of the substrate promiscuity for glycosyltransferases. ACS Synth. Biol. 2019, 8(12), 2718-2725. | PDF
61. Duan, X.#, Ma, X.#, Li, S., and Zhou, Y.* Free fatty acids promote transformation eficiency of yeast. FEMS Yeast Res. 2019, 19(7), foz069. | PDF
2018
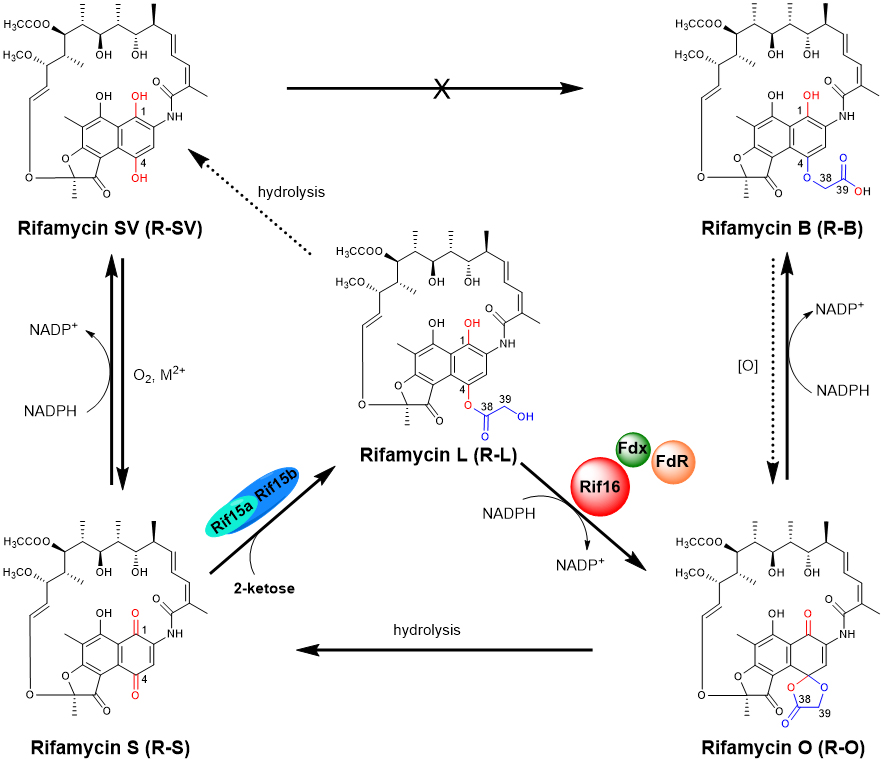
59. Qi, F. #, Lei, C. #, Li, F., Zhang, X., Wang, J., Zhang, W., Fan, Z., Li, W., Tang, G., Xiao, Y.*, Zhao, G., Li, S. * Deciphering the late steps of rifamycin biosynthesis. Nat. Commun. 2018, 9, 2342. | PDF
58. Chen, H., Meng, X., Xu, X., Liu, W., Li, S. * The molecular basis for lipase stereoselectivity. Appl. Microbiol. Biotechnol. 2018, 102(8), 3487-3495. | PDF
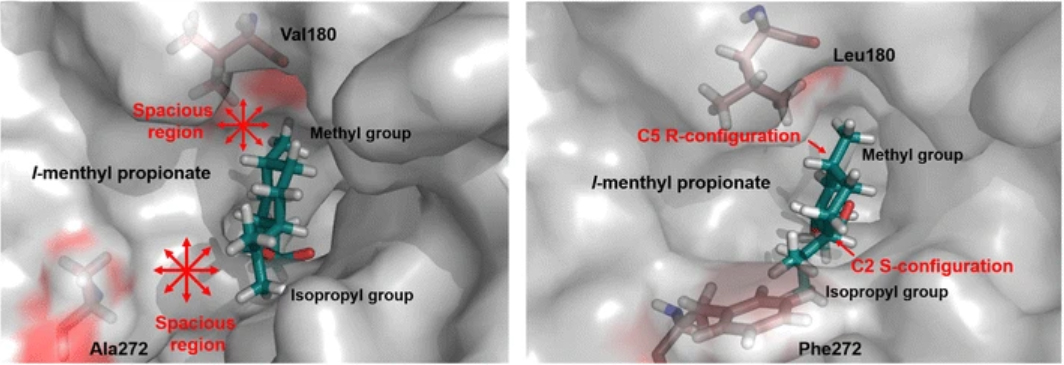
57. Jiang, Y., Li, S.* The catalytic function and application of cytochrome P450 enzymes in biosynthesis and organic synthesis. Chin. J. Org. Chem. 2018, 38, 2307-2323. | PDF
56. Zhang, X. #, Li, P. #, Qin, G., Li, S., Voogd, J. N., Tang, X.*, and Li, G.* Isolation and Absolute Con?gurations of Diversiform C17, C21 and C25 Terpenoids from the Marine Sponge Cacospongia sp. Mar. Drugs 2018, 17(1), 14. | PDF
55. Zhong,.B., Du, L., Li, Z., Zhang, W., Zhang, X., Li, J., Huang, S., Hu, X.*, Li, S., Liu, K.* Biosynthesis of Gastrodin Based on the Cytochrome P450-Mediated Oxidative Biodegradation of 4-Cresol. J. Hunan Normal Univ. (Natural Science) 2018, 41(4), 73-80. | PDF
54. Li, Q.*, Ding, W., Yao, Z., Tu, J., Wang, L., Huang, H., Li, S., and Ju, J.* AbmV Catalyzes Tandem Ether Installation and Hydroxylation during Neoabyssomicin/Abyssomicin Biosynthesis. Org. Lett. 2018, 20, 4854-4857. | PDF
53. Huang, Y., Zheng, X., Pilgaard, B., Holck, J., Muschiol, J., Li, S., and Lange, L.* Identification and characterization of GH11 xylanase and GH43 xylosidase from the chytridiomycetous fungus, Rhizophlyctis rosea. Appl. Microbiol. Biotechnol. 2018, 103(2), 777-791. | PDF
52. Dai, Z., Wang, Y., Zhou, Z., Li, S., Zhang, X *. Synthetic Biology for Production of Plant-derived Natural Products. Bull. Chin. Acad. Sci. 2018, 33(11), 1228-1238. | PDF
2017
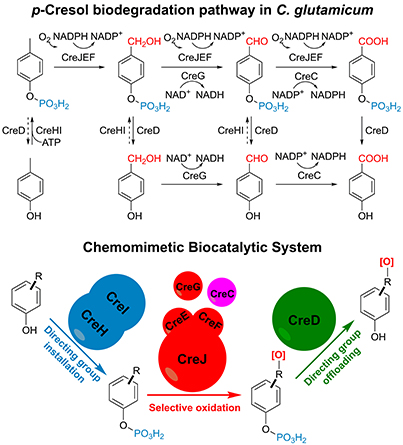
50. Du, L. #, Dong, S. #, Zhang, X., Jiang, C., Chen, J., Yao, L., Wang, X., Wan, X., Liu, X., Wang, X., Huang, S., Cui, Q., Feng, Y.*, Liu, S.*, and Li, S.* Selective oxidation of aliphatic C–H bonds in alkylphenols by a chemomimetic biocatalytic system. Proc. Natl. Acad. Sci. U.S.A. 2017, 114(26), E5129-E5137. | PDF
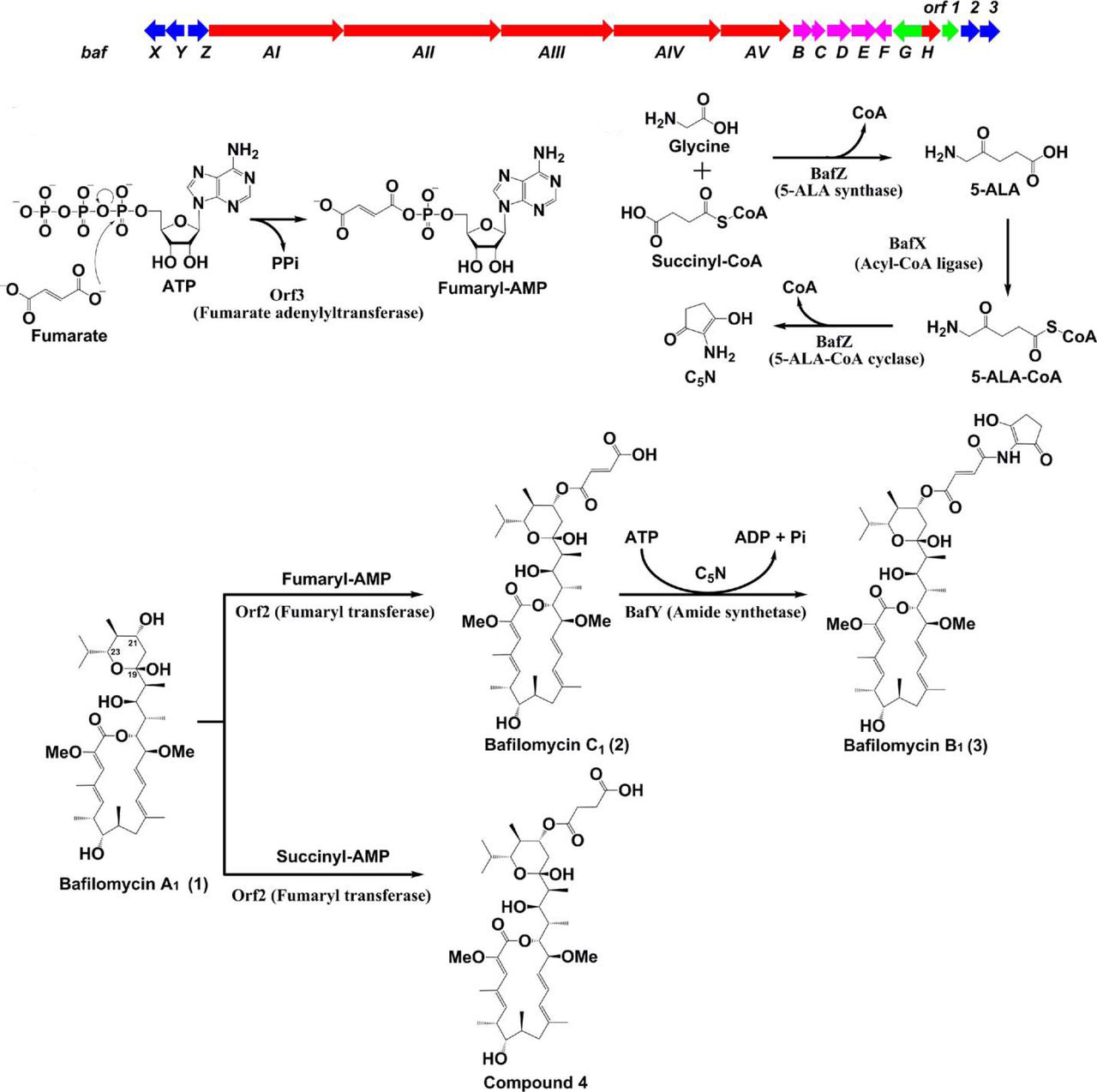
49. Li, Z., Du, L., Zhang, W., Zhang, X., Jiang, Y., Liu, K., Men, P., Xu, H., Fortman, J. L., Sherman, D. H., Yu, B., Gao, S., Li, S.* Complete elucidation of the late steps of bafilomycin biosynthesis in Streptomyces lohii. J. Biol. Chem. 2017, 292(17), 7095-7104. | PDF
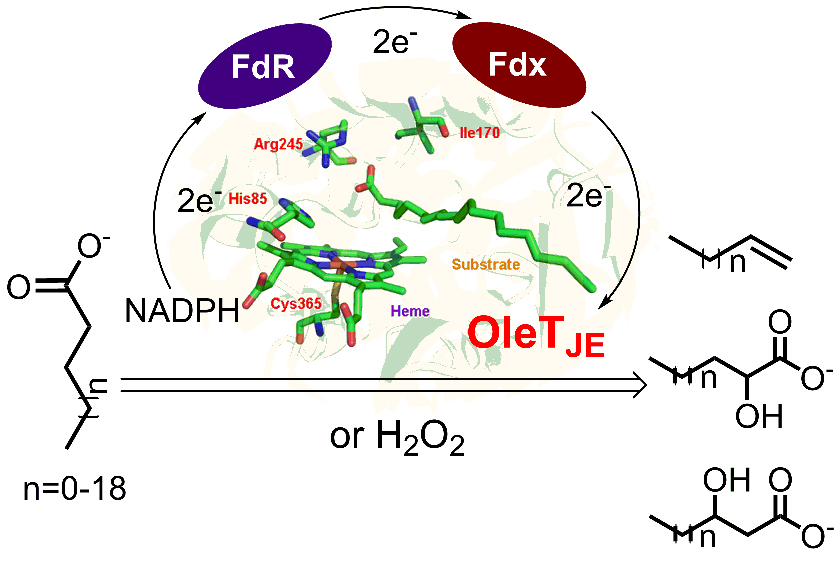
48. Fang, B. #, Xu, H. #, Liu, Y., Qi, F., Zhang, W., Chen, H., Wang, C., Wang, Y., Yang, W., and Li, S.* Mutagenesis and redox partners analysis of the P450 fatty acid decarboxylase OleTJE. Sci. Rep. 2017, 7, 44258. | PDF
47. Xu, H., Ning, L., Yang, W., Fang, B., Wang, C., Wang, Y., Xu, J., Colin, S., Laeuffer, F., Fourage, L., and Li, S. * In vitro oxidative decarboxylation of free fatty acids to terminal alkenes by two new P450 peroxygenases. Biotechnol. Biofuels 2017, 10, 208. | PDF
46. Sun, Y., Ma, L., Chen, H., Xu, H., Zheng, X., Qi, F., Li, S.* Production of α-alkenes catalyzed by the fused lipase and P450 fatty acid de-carboxylase. Chin. Sci. Bull. 2017, 62(1), 1-7. | PDF
45. Yao, Q. #, Ma, Li. #, Liu, L., Ikeda, H., Fushinobu, S., Li, S., and Xu, L.* Hydroxylation of Compactin (ML-236B) by CYP105D7 (SAV_7469) from Streptomyces avermitilis. J. Microbiol. Biotechnol. 2017, 27(5), 956–964. | PDF
44. Zhang, W. #, Huffman, J. #, Li, S., Shen, Y.*, and Du, L.,* Unusual acylation of chloramphenicol in Lysobacter enzymogenes, a biocontrol agent with intrinsic resistance to multiple antibiotics. BMC Biol. 2017, 17, 59-66. | PDF
43. Zhang, W., Zhao, B., Du, L.*, Shen, Y.* Cytotoxic Polyketides with an Oxygen-Bridged Cyclooctadiene Core Skeleton from the Mangrove Endophytic Fungus Phomosis sp. A818. Molecules. 2017, 22, 1547. | PDF
2016
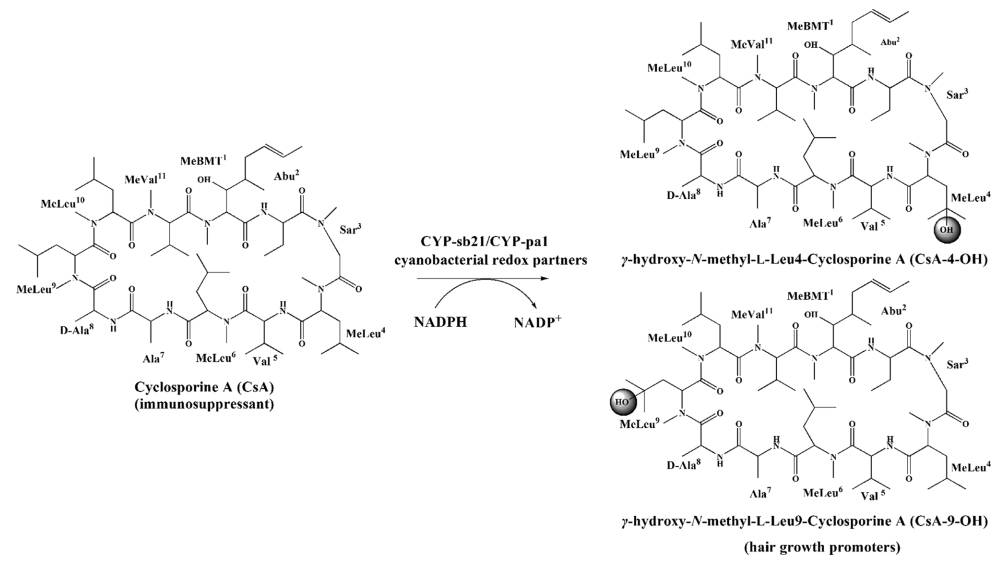
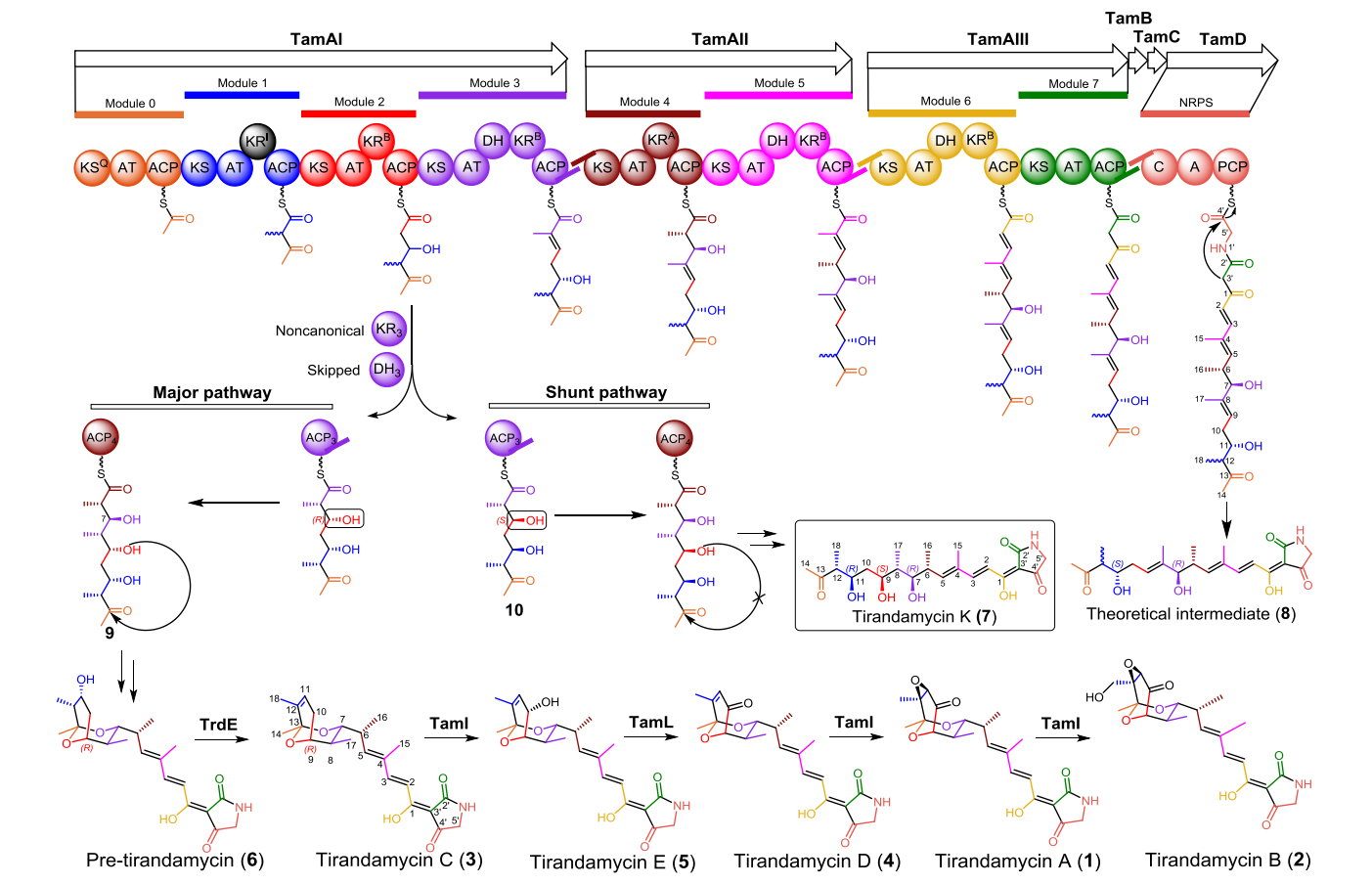
41. Zhang, X., Li, Z., Du, L., Chlipala, G. E., Lopez, P. C., Zhang, W., Sherman, D. H.*, and Li, S.* Identification of an unexpected shunt pathway product provides new insights into tirandamycin biosynthesis. Tetrahedron Lett. 2016, 57(52), 5919-5923. | PDF
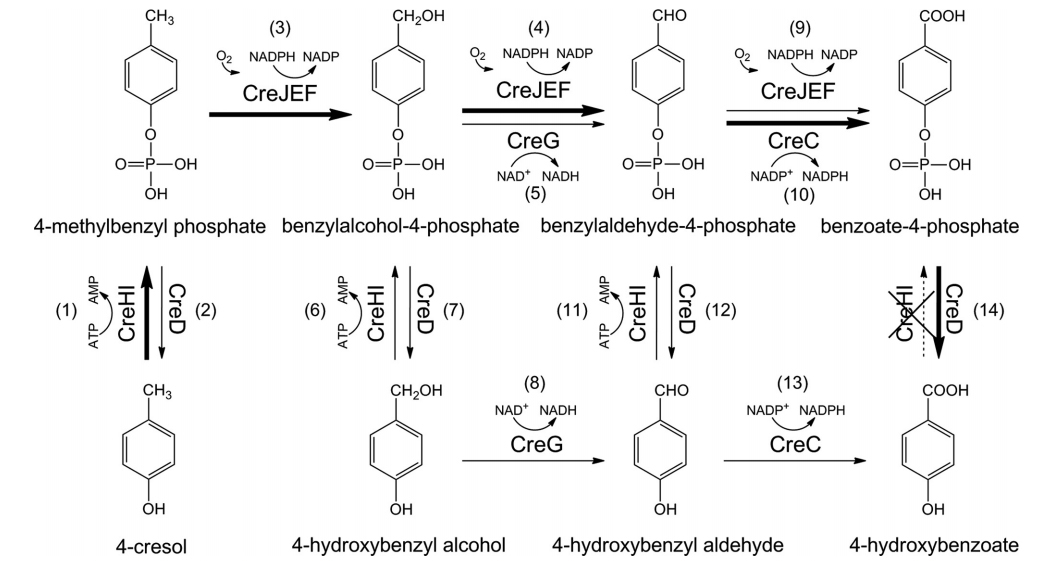
40. Du, L., Ma, L., Qi, F., Zheng, X., Jiang, C., Li, A., Wan, X., Liu, S.-J.*, and Li, S.* Characterization of a Unique Pathway for 4-Cresol Catabolism Initiated by Phosphorylation in Corynebacterium glutamicum. J. Biol. Chem. 2016, 291(12), 6583-6594. | PDF
39. Zheng, X., Fang, B., Han, D., Yang, W., Qi, F., Chen, H., and Li, S.* Improving the Secretory Expression of an α-Galactosidase from Aspergillus niger in Pichia pastoris. PLoS ONE 2016, 11(8), e0161529. | PDF
38. Li, Z., Zhang, W., and Li, S.* Cytochrome P450 enzymes and microbial drug development – A review. Acta Microbiol. Sin. 2016, 56(3), 496-515. | PDF
37. Sun, J., Liu, C., Li, R., Zhang, W. *, Li, S., Functional characterization of PbPT, a prenyltransferase from Penicillium Brevicompactum NRRL 864. J. Qingdao Univ. (E&T), 2016, 31(2),107-115. | PDF
2015
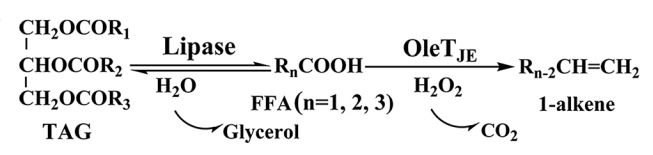
35. Yan, J.*, Liu, Y., Wang, C., Han, B., and Li, S.*Assembly of lipase and P450 fatty acid decarboxylase to constitute a novel biosynthetic pathway for production of 1-alkenes from renewable triacylglycerols and oils. Biotechnol. Biofuels 2015, 8, 34. | PDF
34. Zhang, W., Cao, S., Qiu, L., Qi, F., Li, Z., Yang, Y., Huang, S., Bai, F., Liu, C., Wan, X.,* and Li, S.* Functional Characterization of MpaG’, the O-Methyltransferase Involved in the Biosynthesis of Mycophenolic Acid. Chembiochem 2015, 16(4), 565-569. | PDF
33. Woo, M. W., Lee, B. R., Nah, H.J., Choi, S. S., Li, S., and Kim, E.-S.* Domain Characterization of Cyclosporin Regio-specific Hydroxylases in Rare Actinomycetes. J. Microbiol. Biotechnol. 2015, 25(10), 1634-1639. | PDF
32. Qi, G. #, Wang, D. #, Yu, L., Tang, X., Chai, G., He, G., Ma, W., Li, S., Kong, Y., Fu, C.*, and Zhou, G.* Metabolic engineering of 2-phenylethanol pathway producing fragrance chemical and reducing lignin in Arabidopsis. Plant Cell Rep. 2015, 34(8), 1331-1342. | PDF
31. Bernard, S. M., Akey, D. L., Tripathi, A., Park, S. R., Konwerski, J. R., Anzai, Y., Li, S., Kato, F., Sherman, D. H., and Smith, J. L.* Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases. ACS Chem. Biol. 2015, 10(5), 1340-1351. | PDF
2014
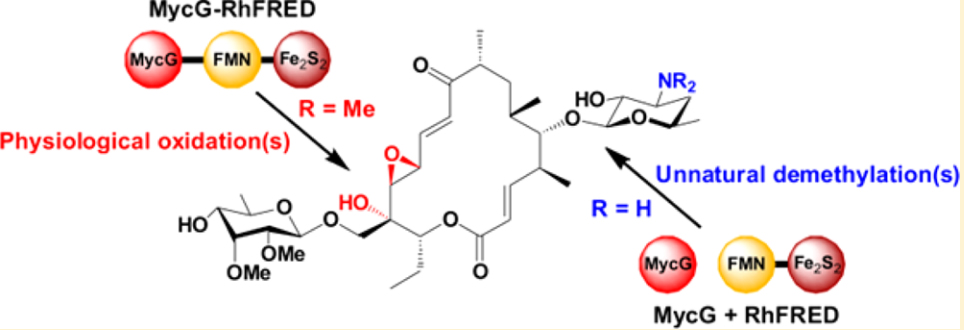
29. Zhang, W., Liu, Y., Yan, J., Cao, S., Bai, F., Yang, Y., Huang, S., Yao, L., Anzai, Y., Kato, F., Podust, L. M., Sherman, D. H.*, and Li, S.* New reactions and products resulting from alternative interactions between the P450 enzyme and redox partners. J. Am. Chem. Soc. 2014, 136(9), 3640-3646. (Faculty of 1000 Prime: http://f1000.com/prime/718276150) | PDF
28. Liu, Y., Wang, C., Yan, J., Zhang, W., Guan, W., Lu, X., and Li, S.* Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol. Biofuels 2014, 7, 28. | PDF
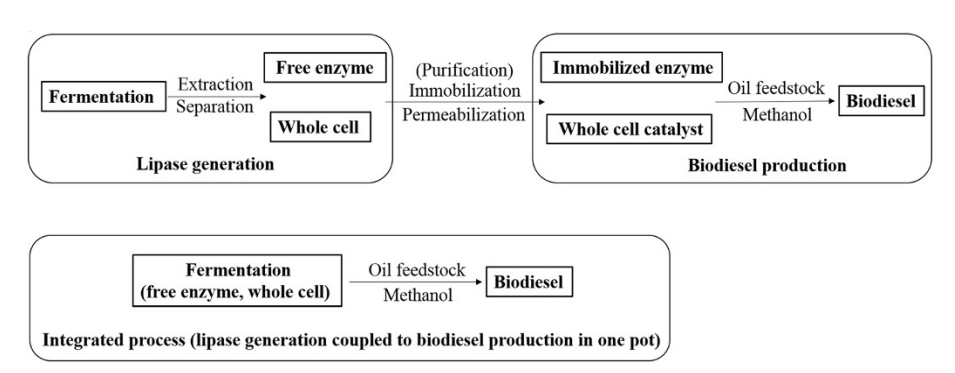
27. Yan, J.*, Zheng, X., and Li, S.* A novel and robust recombinant Pichia pastoris yeast whole cell biocatalyst with intracellular overexpression of a Thermomyces lanuginosus lipase: preparation, characterization and application in biodiesel production. Bioresour. Technol. 2014, 151, 43-48. | PDF
26. Zhang, W. #, Xu, L. #, Yang, L., Huang, Y., Li, S., and Shen. Y.* Phomopsidone A, a novel depsidone metabolite from the mangrove endophytic fungus Phomopsis sp. A123. Fitoterapia 2014, 96, 146-151. | PDF
25. Huang, S.*, Gao, J., Wu, R., Li, S. and Bai Z.* Polydimethylsiloxane: A General Matrix for High-Performance Chromatographic NMR Spectroscopy. Angew. Chem. Intl. Ed. 2014, 53, 11592-11595. | PDF
24. Wang, Q. #, Huang, X. #, Zhang, J., Lu, X., Li, S., and Li, J.-J.* Engineering self-sufficient aldehyde deformylating oxygenase fused to alternative electron transfer systems for efficient conversion of aldehydes into alkanes. Chem. Comm. 2014, 50, 4299-4301. | PDF
2013
22. Sunderhaus, J. D., McAfoos, T. J., Finefield, J. M., Kato, H., Li, S., Tsukamoto, S., Sherman, D. H., and Williams, R. M.* Synthesis and bioconversions of notoamide T: a biosynthetic precursor to stephacidin A and notoamide B. Org. Lett. 2013, 15(1), 22-25. | PDF
2012
20. Li, S., Anand, K., Tran, H., Yu, F., Finefield, J. M., Sunderhaus, J. D., McAfoos, T. J., Tsukamoto, S., Williams, R. M.*, and Sherman, D. H.* Comparative analysis of the biosynthetic systems for fungal bicyclo[2.2.2]diazaoctane indole alkaloids: the (+)/(-)-notoamide, paraherquamide and malbrancheamide pathways. MedChemComm 2012, 3(8), 987-996. | PDF
19. Li, S., Finefield, J. M., Sunderhaus, J. D., McAfoos, T. J., Williams, R. M.*, and Sherman, D. H.* Biochemical characterization of NotB as an FAD-dependent oxidase in the biosynthesis of notoamide indole alkaloids. J. Am. Chem. Soc. 2012, 134(2), 788-791. | PDF
18. Anzai, Y.*, Tsukada, S. I., Sakai, A., Masuda, R., Harada, C., Domeki, A., Li, S., Kinoshita, K., Sherman, D. H., and Kato, F. Function of the cytochrome P450 enzymes MycCI and MycG in Micromonospora griseorubida, a producer of the macrolide antibiotic mycinamicin. Antimicrob. Agents Chemother. 2012, 56(7), 3648-3656. | PDF
2011
16. Akey, D. L., Li, S., Konwerski, J., Confer, L., Bernard, S. M., Anzai, Y., Kato, F., Sherman, D. H., and Smith, J. A.* New structural form in the SAM/metal-dependent O-methyltransferase family: MycE from the mycinamycin biosynthetic pathway. J. Mol. Biol. 2011, 413(2), 438-450. | PDF
2010
14. McAfoos, T. J., Li, S., Tsukamoto, S., Sherman, D. H., and Williams, R. M.* Studies on the Biosynthesis of the stephacidins and notoamides. Total synthesis of notoamides. Heterocycles 2010, 82(1), 461-472. | PDF
13. Tsukada, S., Anzai, Y., Li, S., Kinoshita, K., Sherman, D. H., and Kato, F.* Gene targeting for O-methyltransferase genes, mycE and mycF, on the chromosome of Micromonospora griseorubida producing mycinamicin with a disruption cassette containing the bacteriophage φC31 attB attachment site. FEMS Microbiol. Lett. 2010, 304(2), 148-156. | PDF
12. Carlson, J. C., Fortman, J. L., Anzai, Y., Li, S., Douglas, A. B., and Sherman, D. H.* Identification of the tirandamycin biosynthetic gene cluster from Streptomyces sp. 307-9. Chembiochem 2010, 11(4), 564-572. | PDF
2009
10. Carlson, J. C. #, Li, S.#, Douglas, A. B., and Sherman, D. H.* Isolation and characterization of tirandamycins from a marine-derived Streptomyces sp. J. Nat. Prod. 2009, 72(11), 2076-2079. (*Co-first authors) | PDF
9. Li, S., Anzai, Y., Kinoshita, K., Kato, F., Sherman, D. H.* Functional analysis of MycE and MycF, two O-methyltransferases involved in the biosynthesis of mycinamicin macrolide antibiotics. Chembiochem 2009, 10(8), 1297-1301. | PDF
8. Li, S., Ouellet, H., Sherman, D. H., Podust, L. M.* Analysis of transient and catalytic desosamine-binding pockets in cytochrome P-450 PikC from Streptomyces venezuelae. J. Biol. Chem. 2009, 284(9), 5723-5730. | PDF
2008
2007
5. Li, S., Grüschow, S., Dordick, J. S., Sherman, D. H.* Molecular analysis of the role of tyrosine 224 in the active site of Streptomyces coelicolor RppA, a bacterial type III polyketide synthase. J. Biol. Chem. 2007, 282(17), 12765-12772. | PDF
2006
2004
2003
1. Li, S., Zheng, Z.* Discovery and development of marine antitumor natural products. Mar. Sci. Bull. 2003, 22(2), 76-82. | PDF
PATENTS
2. Li Shengying, David H. Sherman, Mani Raj Chaulagain, John Montgomery, and Allison R. Knauff. A Method for Selective Oxidation of C-H Bonds by an Engineered P450 Enzyme. (WO 2011038313A2)
3. Li Shengying, David H. Sherman, Krithika Anand, and Robert M. Williams. Biosynthetic Systems Producing Fungal Indole Alkaloids. (US 9650656 B2)
4. Yan, Jinyong, Li Shengying. The invention relates to a method for catalyzing the production of biodiesel by coupling lipase production.(201410138507.3)
5. Li Shengying, Liu Yi, Lv Xuefeng, and Liu Xufeng. The invention relates to a method for producing terminal olefins and its application.(201410660346.4)
6. Yan Jinyong, Li Shengying. The invention relates to aliphatic olefin catalytic synthesis method based on coupling catalysis of lipase and P450 fatty acid decarboxylase.(ZL201510003451.5)
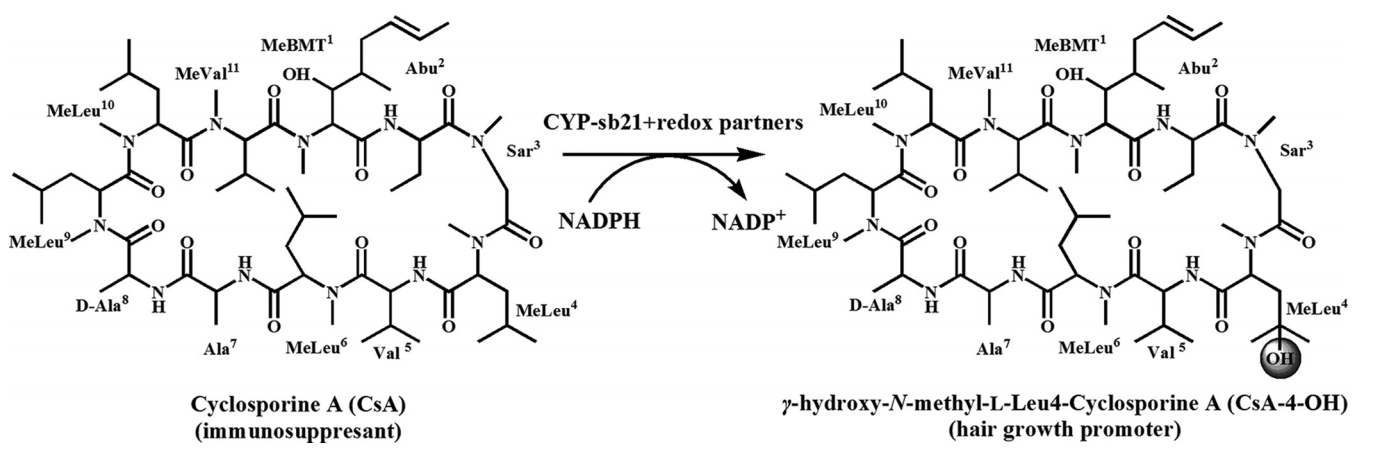
7. Li Shengying, Ma Li, and Kim Eung-Soo. The invention relates to a biological transformation method for producing hair growth promoter.(201510346350.8)
8. Li Shengying, Du lei, and Liu Shuangjiang. The invention relates to a catalytic oxidation system of phenolic compounds and its application.(201610015698.3)
9. Li Shengying, Zheng Xianliang. The construction and application of engineering bacteria with high secretion and expression of Aspergillus Niger alpha-galactosidase AGA.(2015107066150)
10. Li Shengying, Fang Bo, Qi Fengxia, Zhang Wei, and Du Lei. The invention relates to combination of redox partners to support the activity of P450 fatty acid decarboxylase and its application.(201610151612.X)
11. Fourage Laurent, Laeuffer Frederie, Strub Henri, Wang Yun, Xu Jian, Xu Huifang, and Li Shengying. Production of Alpha-Olefins. (WO 2017001606A1)
12. Li Shengying, Xu Huifang, and Ning Linlin. Method for detecting hydrogen peroxide conversion activity of an enzyme. ( EP17290115.9)
13. Li Shengying, Xu Huifang, and Ning Linlin. Improved cytochrome P450 fatty acid decarboxylases. ( EP18290089.4)
14. Li Shengying, Li Zhong, Du Lei, Zhang Xingwang, and Zhang Wei. The invention relates to the construction of an engineering bacteria with high yield of pavloamycin and its application.(201811634377.7)